This fun project was driven by the talented and rigorous Alex Gillis @cftbla.bsky.social, in wonderful collaboration with Ana Tufegdžić Vidaković's @anatv.bsky.social lab!
@singmolsci.bsky.social
@unswrna.bsky.social
15.01.2026 05:25 — 👍 4 🔁 0 💬 0 📌 0

🎉 𝙒𝙝𝙖𝙩 𝙖 𝙙𝙖𝙮! 🎉
On Thursday we held our inaugural EMCR Mini Symposium, bringing together six in-person nodes plus online attendees from across Australasia. It was fantastic to see our community come together to share science, build connections, and finish 2025 strong.
#EMCR #AEpiA #epigenetics
05.12.2025 22:48 — 👍 0 🔁 1 💬 1 📌 0
Oh! Would have been great to see you (I live in Wollongong now). But I am off to Korea tomorrow! Enjoy the gong. Hope the weather improves. 🤞
29.10.2025 06:16 — 👍 1 🔁 0 💬 1 📌 0

🌟 𝐈𝐧𝐭𝐫𝐨𝐝𝐮𝐜𝐢𝐧𝐠 𝐄𝐌𝐂𝐑 𝐌𝐢𝐧𝐢 𝐒𝐲𝐦𝐩𝐨𝐬𝐢𝐮𝐦 𝐂𝐚𝐫𝐞𝐞𝐫 𝐏𝐚𝐧𝐞𝐥𝐥𝐢𝐬𝐭 – 𝐃𝐫 𝐋𝐮𝐤𝐞 𝐈𝐬𝐛𝐞𝐥 🌟
We’re excited to spotlight Dr Luke Isbel, one of our inspiring career development panellists for the upcoming Australasian Epigenetics Alliance 𝐄𝐌𝐂𝐑 𝐌𝐢𝐧𝐢 𝐒𝐲𝐦𝐩𝐨𝐬𝐢𝐮𝐦!
aepia.org.au/emcr-symposium
09.10.2025 22:25 — 👍 3 🔁 3 💬 1 📌 0
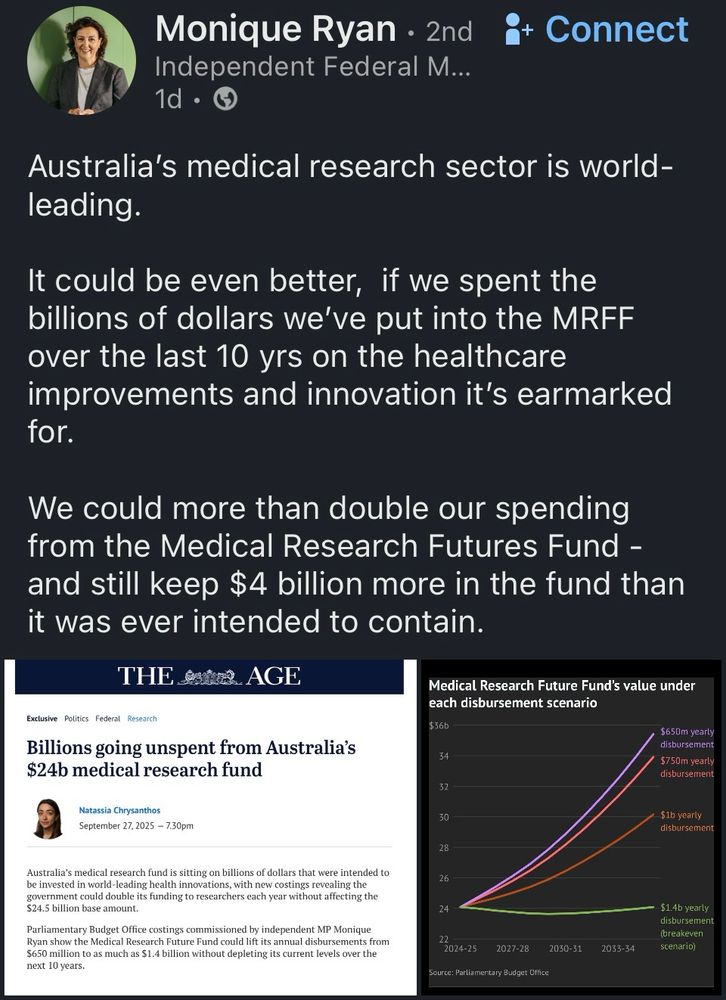

A purpose built solution to Australia’s biomedical funding crisis….. release the #MRFF = invest in our future scientists & their discoveries. #DiscoveriesNeedDollars www.smh.com.au/politics/fed...
29.09.2025 01:51 — 👍 13 🔁 9 💬 1 📌 0

The Asia RNA Club Symposium will be happening in Seoul in Nov 3-5, bringing together RNA scientists from the Asia-Pacific region. Excellent speakers and a great opportunity to connect! Abstract submission by Sept 5 asiarnaclub.org
21.08.2025 08:04 — 👍 2 🔁 3 💬 1 📌 0
Great list! Can I be added too please?
28.08.2025 21:12 — 👍 0 🔁 0 💬 1 📌 0

Research
The immune system is a spatially distributed, whole-body sensory-reactive system that monitors, reacts, and adapts to various stress signals (e.g., tissue
We have some news to share: The Kueh lab is moving to Yale this fall! We will be joining the Yale Center for Systems and Engineering Immunology @yalecsei.bsky.social and the Department of Immunobiology.
medicine.yale.edu/systems-engi...
19.08.2025 02:53 — 👍 25 🔁 3 💬 2 📌 1
Fantastic to see this short film about A/Prof Lawrence Lee who is a Group Leader in UNSW Single Molecule Science and the founding CEO of the exciting startup, SWAN Genomics 🧬🔬
17.05.2025 23:09 — 👍 6 🔁 2 💬 0 📌 0
Research Assistant / Research Associate (Fixed Term) - Job Opportunities - University of Cambridge
Research Assistant / Research Associate (Fixed Term) in the Department of Plant Sciences at the University of Cambridge.
Join our group (www.luginbuehllab.com) at @camplantsci.bsky.social! Are you excited about mycorrhiza, (single-cell) transcriptomics, carbon physiology, and the model crop rice? We have a 2-year postdoc position available, starting ASAP. Closing date 23rd May 2025!
www.jobs.cam.ac.uk/job/51214/
04.05.2025 11:13 — 👍 26 🔁 25 💬 0 📌 4
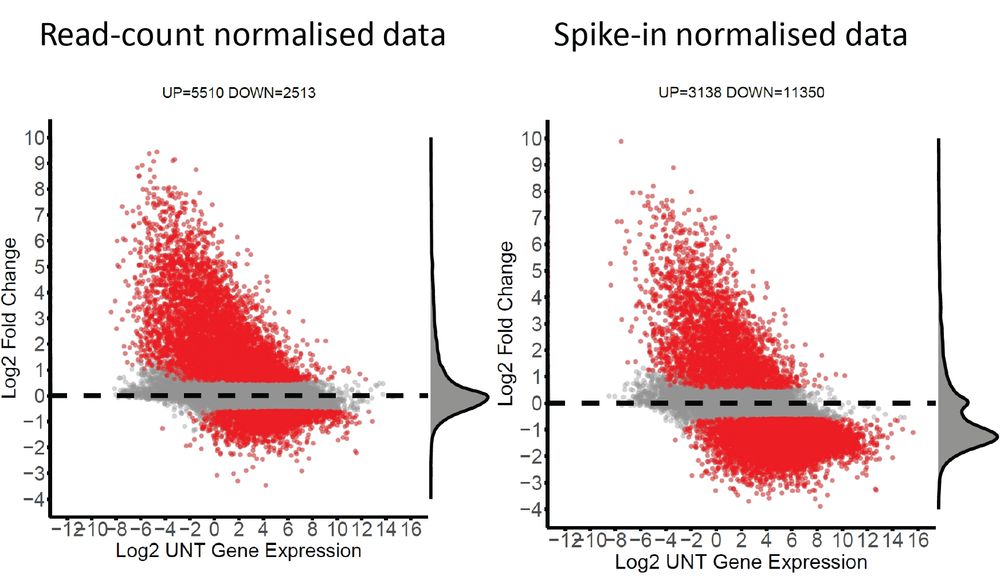
We have had multiple discussions about why people have not previously observed the very dramatic reduction in transcription upon PNUTS depletion. So I looked at our own TT-seq data and analysed it without including the spike-in. The numbers speak for themselves! Spike-in calibration is important!
12.12.2024 10:49 — 👍 27 🔁 10 💬 1 📌 2

Cartoon showing two people working at a conveyer belt which is processing Pol II complexes. Aberrant Pol II complexes are being removed from the conveyer belt and placed in recycling or rubbish bins. This represents the checkpoint formed by CRL3ARMC5 ubiquitin ligase and Integrator phosphatase which controls the quality and quantity of transcription complexes on genes.
Mechanism of quality control of gene expression revealed by @anatv.bsky.social & @scottbscience.bsky.social’s groups in study of Pol II molecules.
Read more about the work, led by Roberta Cacioppo, Alexander Gillis, Iván Shlamovitz & Andrew Zeller: www2.mrc-lmb.cam.ac.uk/newly-identi...
#LMBResearch
11.12.2024 16:05 — 👍 2 🔁 2 💬 0 📌 0
Was a pleasure working with @anatv.bsky.social on this!
12.12.2024 05:39 — 👍 4 🔁 2 💬 0 📌 0
Thanks to support from @singmolsci.bsky.social @unswrna.bsky.social
12.12.2024 05:38 — 👍 1 🔁 0 💬 1 📌 0

When both ARMC5 and INTS8 are depleted, excess Pol II is released into genes but often fails to transcribe the full transcription unit, resulting in major gene expression changes. There is lots more in the paper, of course!
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0

This finding pushed us to try to understand what stops Pol II from entering the elongation phase and actually making more RNA. We found that ARMC5 and Integrator phosphatase play complementary roles in this process.
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0

ARMC5 also regulates Pol II levels during normal cell growth - seemingly responsible for around half the cell’s Pol II turnover. But the extra Pol II that accumulates in cells upon ARMC5 loss doesn’t lead to more Pol II in gene bodies: it is mostly in the free pool or in the proximal promoter.
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0

However, using live-cell imaging and cellular fractionation, we found that this excess Pol II mostly accumulates off-chromatin.
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0

It has long been known that Pol II is degraded when transcription is inhibited - either with small molecules or if specific cofactors are depleted. We now know the ubiquitin ligase responsible is CUL3-ARMC5. When we remove ARMC5, Pol II is no longer lost upon inhibition.
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0
Building on our previous work on RNA concentration homeostasis doi.org/10.1016/j.ce..., we were interested to understand how global levels of Pol II are controlled. We teamed up with @anatv.bsky.social's lab who delve deep into mechanisms of ubiquitin-mediated regulation of the transcription cycle.
12.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0
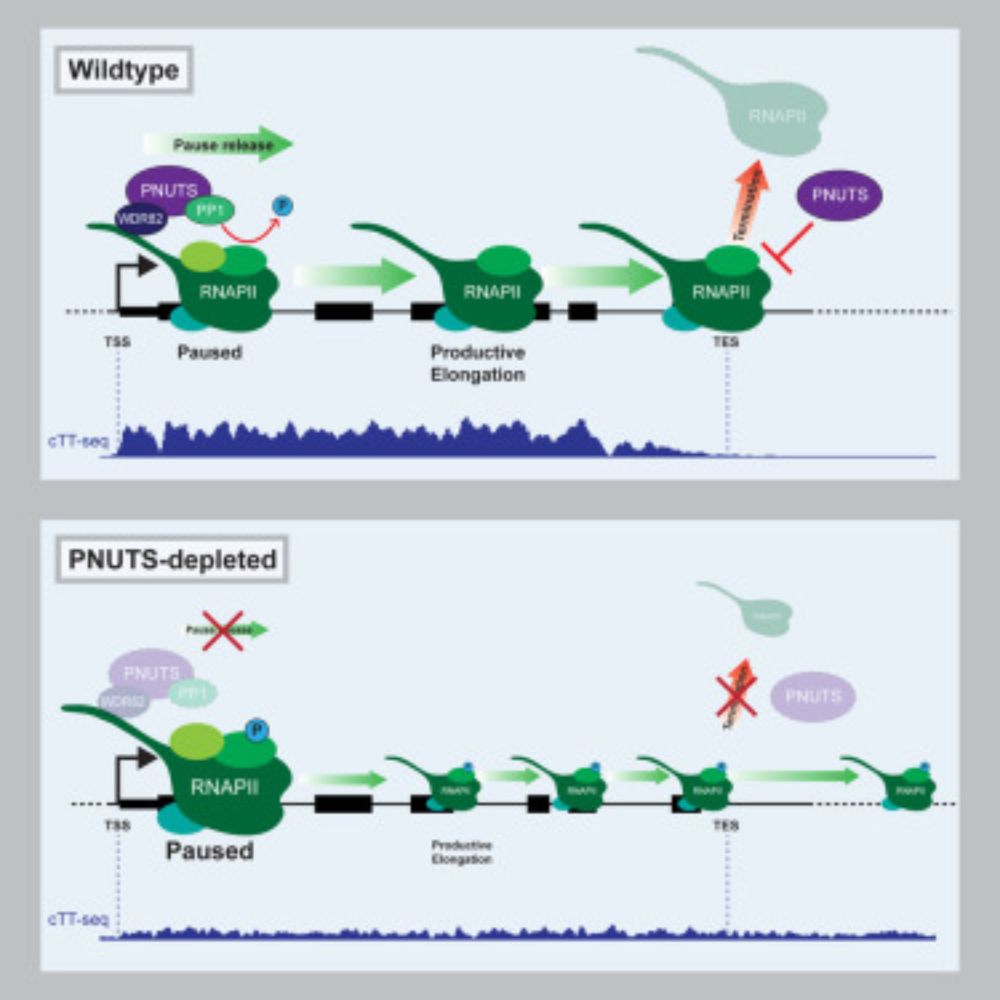
The PNUTS phosphatase complex controls transcription pause release
Kelley et al. discover that the PNUTS phosphatase complex plays an essential role
in gene transcription by controlling RNA polymerase II pause release. PNUTS achieves
this through its TND, which recog...
Very excited that our paper describing a critical role for the PNUTS-PP1 phosphatase complex in transcription pause release is up online! This was a massive team effort with @edimitrova.bsky.social and the rest of the @robklose.bsky.social lab www.cell.com/molecular-ce... A short thread: (1/8)
26.11.2024 16:18 — 👍 66 🔁 28 💬 2 📌 4
The Sydney Protein Group is made up of protein scientists from academia, hospitals and industry.
https://www.asbmb.org.au/sydney-protein-group
Just another LLM. Tweets do not necessarily reflect the views of people in my lab or even my own views last week. http://rajlab.seas.upenn.edu https://rajlaboratory.blogspot.com
Imaging transcription to uncover mechanisms of gene regulation
https://www.rdm.ox.ac.uk/people/edward-tunnacliffe
Stanford BioE, Genetics & Sarafan ChEM-H. Chan-Zuckerberg Biohub Investigator. Our lab develops and applies microfluidic assays for high-throughput biophysics and biochemistry.
Associate Professor / enhancers - 3D genome - morphogenesis
Website: https://www.unige.ch/medecine/gede/en/research-groups/999andrey
Incoming Associate Professor @ Colorado University Anschutz | cancer epigenetics | DNA replication | protein synthesis | vanrechemlab.com
A bit of 3D gene regulation, single-cell omics and transgenic models.
Born and raised in the bay of Algeciras. Enjoying Science and Flamenco at CABD, Seville.
Lab website: https://lupianezlab.github.io/Website/
Email: dario.lupianez@csic.es
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
Scientist in the School of Biotechnology and Biomolecular Sciences (BABS) at UNSW Sydney studying gene regulation and transcription factors in adipose tissue-resident immune cells and red blood cells
Deputy Director at WEHI, Melbourne Australia.
Lab head studying epigenetic control, in the context of X inactivation, genomic imprinting, SMCHD1, Prader Willi Syndrome, FSHD.
Mum, wife, beach lover.
Lecturer, researcher in CRISPR gene editing, epigenetics etc, and university administrator
Computational & Genomics Lab studying 3D genome function and dynamics @ Netherlands Cancer Institute
Enhancers, 3D genome organisation, pluripotent stem cells
Babraham Institute and Enhanc3D Genomics
Professor for Genetics interested in chromatin structure, histone variants and histone modifications and their roles in gene regulation and disease development.
Group leader at the Danish Cancer Institute and Professor at University of Copenhagen #NNFCPR. Interested in epigenetics, genome maintenance, aging & cancer, creativity & innovation, society in general. Opinions are my own.
Bloomberg Distinguished Professor of Structural Biophysics and Chromatin Biology at Johns Hopkins University, #Transcription #DNArepair #Chromatin #cryoEM
Director of Institut Curie research center
Group leader, X chromosome lover, Epigenetics, Development, ncRNA
Cardiac developmental biologist. Oakland and San Francisco. Hi friends! 🇨🇦
Interested in how cells use genes to create shape, form and function in #embryos as well as in Societal Wellbeing
#NotInTheGenes #InNumbersWeTrust #gastruloids #gastrulation