All this work was done in collaboration with @bastien-boussau.bsky.social, Nicolas Lartillot and @laurentjacob.bsky.social.
Relevant links
preprint: arxiv.org/abs/2510.12976
repo: gitlab.in2p3.fr/deelogeny/wp...
If you find this interesting, please share, and we welcome your feedback!
17/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 0 📌 0

A plot showing the frequencies of leaf-bipartitions in trees sampled with an MCMC method (x-axis) and trees sampled with PF2 (y-axis). Overall, if a split is not frequent in MCMC samples, it will also not be very frequent in PF2 samples. The opposite is also true. MCMC samples are bimodal, making it hard to assess calibration visually.
Compared to samples from RevBayes, an MCMC method, we can see that PF2 is broadly in agreement albeit with a smoother distribution. Since PF2 is amortized, it can sample from the posterior much faster than MCMC methods.
16/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

Topological reconstruction accuracy for the same methods under the CherryML model, which accounts for co-evolution between sites, and the SelReg model that takes into account selection pressures. In both cases, PF2 outperforms PF1 by a significant margin, and maximum-likelihood methods by an even bigger one.
Under complex evolution models (e.g. accounting for co-evolution or selection) where the likelihood is not computable, PF2 really shines further improving on the PF1 performance gain over maximum-likelihood methods.
15/17
16.10.2025 20:34 — 👍 2 🔁 0 💬 1 📌 0

A figure describing the runtime and memory usage of the tree-inference methods. PF2 with only topology is the fastest method, the full PF2 with branch lengths is slower than PF1. Both PF1 and PF2 have similar memory usage once the trees get bigger than 50 leaves.
PF2 has a smaller memory footprint than PF1, despite having more parameters. It can run an order of magnitude faster than even FastME when only sampling topologies.
14/17
16.10.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0

Topological reconstruction accuracy of various tree-inference methods on a test set. PF2 performs much better than any other method for trees from 10 to 140 leaves.
We trained 2 PF2 models under LG, a simple model where maximum-likelihood methods shine. PF2 estimates full tree posteriors, while PF2topo only estimates the posterior of the tree topology.
Both models outperform all other methods significantly for smaller trees.
13/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

The method to sample a merge from the learned distribution. PF2 computes the softmin over the sequence pair representations and uses those values as merge smapling weights. PF2 also computes parameters for the Gamma and Beta distribution from which to sample branch-lengths.
At inference time, we can sample trees by iteratively sampling merges and branch lengths from the learned conditional merge and branch-length distributions.
12/17
16.10.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0

The method to compute a single-merge probability from the sequence embeddings. The topological merge probability is parametrized by a softmin over pairs of sequence representations. Branch length probabilities are parametrized by a Gamma and a Beta distribution, whose parameters are also derived from sequence embeddings.
Our probability estimation network, BayesNJ, is also parametrized and learnable. It is jointly optimized with the embedding network EvoPF during training.
11/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0
To estimate the posterior, we describe trees as an ordered succession of pairwise merges. Allowing us to factorize the total tree probability as the product of successive conditional merge probabilities.
10/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

The architecture of the EvoPF embedding network. It shows how a sequence representation and sequence pair representation are each updated using self-attention and mutual-update operations.
Our embedding network, EvoPF, is inspired from EvoFormer and keeps mutually updating representations of sequences and sequence pairs. It is much more scalable than PF1, allowing us to us 250x more learnable parameters than PF1 with a similar memory footprint.
9/17
16.10.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0

At inference time, PF2 embeds an alignment into the learned latent space. PF2 can then sample trees from the estimated posterior distribution using the latent representation.
Once trained, we can embed empirical sequences into the learned latent space and sample trees from the learned posterior, without ever having to compute a likelihood.
8/17
16.10.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0

During training, tree and sequence alignment pairs are sampled from a simulator under a chosen probabilistic model, this defines our prior distribution. PF2 then embeds the sequence alignment into a latent space using the EvoPF embedding module. The BayesNJ module is then used to produce an estimate of the posterior probability of the tree given the latent representation of the sequence alignment. The negative log of this estimated is then minimized during training to find the optimal set of PF2 parameters.
PF2 estimates the posterior probability of a tree by (1) embedding sequences into a latent space, then (2) estimating the posterior from that latent space.
PF2 is optimized by minimizing the estimated log-posterior over many (tree, sequences) pairs sampled under a given probabilistic model.
7/17
16.10.2025 20:34 — 👍 1 🔁 0 💬 1 📌 0
To solve this, we initially thought of learning through a neighbor joining step. This proved complicated. Thankfully, there is a way that does not require differentiating through discrete steps: Neural Posterior Estimation (NPE).
6/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

A plot showing the topological accuracy of different methods on a test set. PF1 performs better than other distance-based methods but worse than maximum-likelihood methods.
Our first attempt, PF1, used pairwise distances as a proxy learning target. This worked reasonably well, but using a proxy yields a gap in topological accuracy w.r.t maximum likelihood methods.
(see doi.org/10.1093/molb...)
5/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

A visual justification of simulation-based and likelihood-free inference: under some probabilistic models, computing likelihoods is hard but sampling data is easy.
Likelihood-free/simulation-based inference can solve this issue.
It allows for inference under complex models where sampling is easy but likelihood computations costly or impossible.
It's an alternative way to access the model.
4/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

A diagram of maximum likelihood based tree inference: the method searches tree space for the tree that maximizes the likelihood of the observed sequence data under a chosen probabilistic model of sequence evolution.
Maximum-likelihood based tree inference searches for the most likely tree over the sequences.
This yields accurate estimates but can be slow and limits us to evolution models for which we can compute the likelihood.
3/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0

A diagram of phylogenetic inference: we build a tree summarizing how a given set of sequences evolved from a common ancestor.
Phylogenetic trees describe the shared evolutionary history of a set of related genomic sequences (at the leaves) from a common ancestor (at the root).
The topology describes this ancestry, while the branch lengths describe some measure of how related or distant sequences at each end are.
2/17
16.10.2025 20:34 — 👍 0 🔁 0 💬 1 📌 0
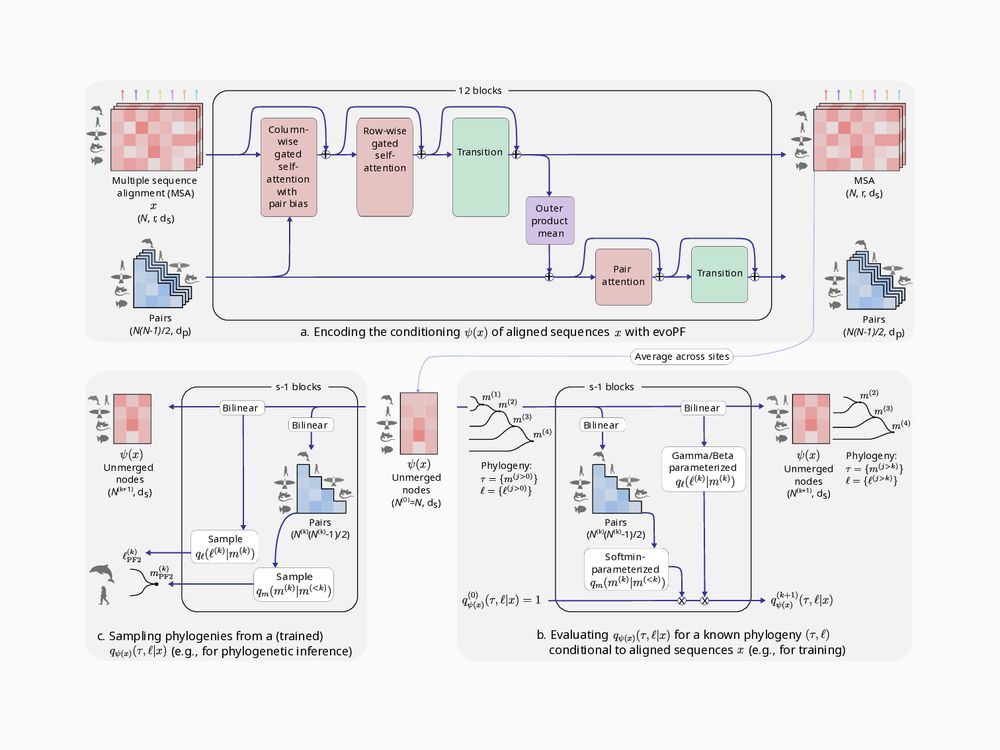
A three panel figure describing the Phyloformer 2 (PF2) method. The top panel describes the architecture of the PF2 neural network. The bottom right panel describes how PF2 estimates the posterior probability of a tree given a sequence emebdding at train time. The bottom left panel describes how PF2 samples trees from the learned posterior at inference time.
We’re very excited to finally share our latest work:
Phyloformer 2, a deep end-to-end phylogenetic reconstruction method: arxiv.org/abs/2510.12976
Using neural posterior estimation, it outperforms Phyloformer 1 and maximum-likelihood methods under simple and complex evolutionary models.
🧵1/17
16.10.2025 20:34 — 👍 7 🔁 2 💬 1 📌 2
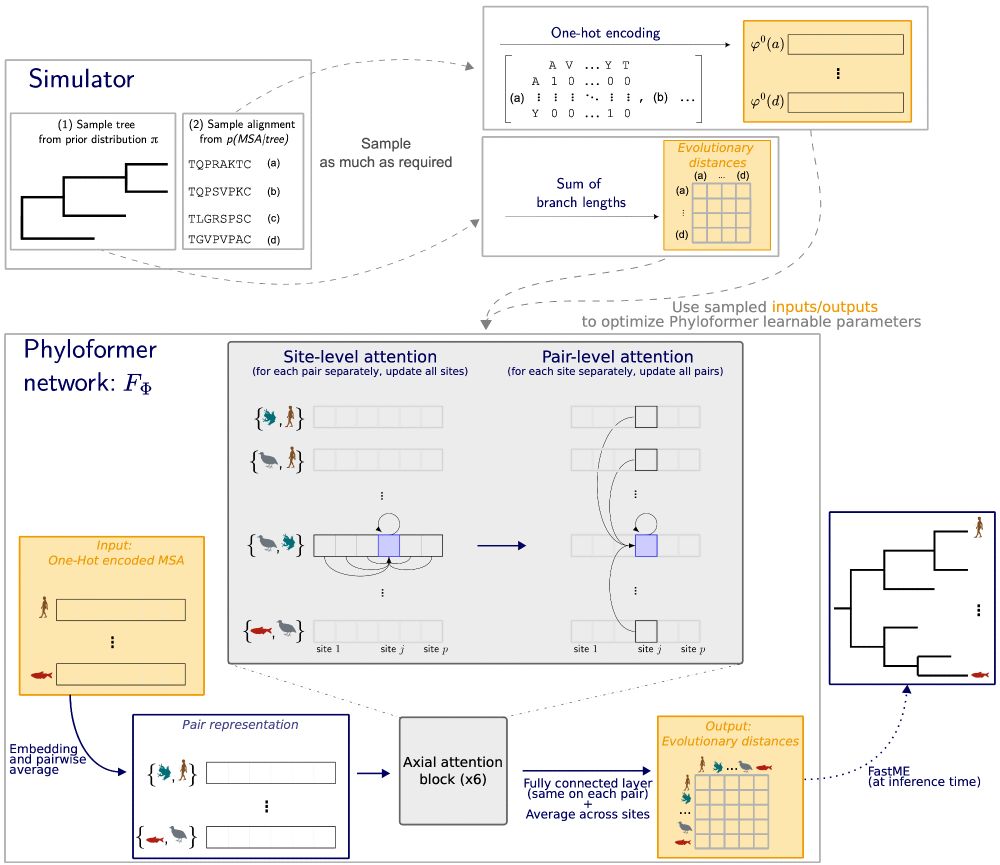
A sketch summarizing the entire Phyloformer process.
All this work was done by Luca Nesterenko and
@lblassel.bsky.social , assisted by P. Veber, Bastien Boussau
and myself.
The code and data are available at github.com/lucanest/Phy...
Please share if you find this interesting, and we welcome your feedback :)
24.06.2024 08:35 — 👍 1 🔁 1 💬 0 📌 0

A logo symbolizing the Phyloformer process: it starts from an artificial neural networks and ends with a phylogeny.
We just released a preprint for Phyloformer, a likelihood-free inference method for phylogenetic reconstruction: biorxiv.org/content/10.1...
Faster than distance methods like neighbor joining, it outperforms maximum likelihood methods under complex models of sequence evolution.
🧵
24.06.2024 08:24 — 👍 3 🔁 2 💬 1 📌 1
Research leader @NHM | Animal evolution | phylogenomics | molecular evolution
Associate Professor of Biological Sciences at University of Pittsburgh.
Adaptive and Convergent Evolution in response to extreme environments. Phylogenetics. Diving mammals. Subterranean life. Visual adaptations.
http://nclarklab.org
Researcher at Uni Cambridge working on bacterial evolution and AI
Research Associate, Imperial College London 🍄 Fungal genomics and bioinformatics
Associate professor at ETH Zurich, studying the cellular consequences of genetic variation. Affiliated with the Swiss Institute of Bioinformatics and a part of the LOOP Zurich.
Researcher in computational biology / bioinformatics at @i2bcparissaclay.bsky.social 🇫🇷
Protein-protein, protein-RNA & protein-DNA interactions, structure & evolution
ML & DL
Biology of genome maintenance
Women/diversity in science
Personal account
CNRS researcher in bioinformatics
Lille, France (Bonsai team).
I develop efficient computational methods to analyze massive sequencing data, creating scalable tools for genomics, transcriptomics, and metagenomics.
https://malfoy.github.io/
interested in algorithmic bioinformatics 💾🧬
Research group in Lille, Fr.
We are the largest biomedical science hub in the south of Europe, by the beach in Barcelona.
Recerca biomèdica amb vistes |
@researchmar.bsky.social @crg.eu @melisupf.bsky.social @embl.org @isglobal.org @ibe-barcelona.bsky.social @fpmaragall.bsky.social
Média indépendant en ligne, sans pub et en accès libre sur l'actualité sociale et écologique, financé par les dons des lecteurs et lectrices.
Notre rythme sur basta.media : un jour, une enquête. Parce que bien s'informer, c'est déjà s'engager.
Sequence bioinfomatician, algorithms, methods.
Postdoc in Institut Pasteur in Rayan Chikhi's lab
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
Research scientist. I like algorithms, bioinformatics, computer science, and HPC.
Biochemist, now working in science policy.
Passionate about biodiversity 🪲🌿, science and innovation policy #scipol, scientific policy advice, science diplomacy and of course the life sciences (cancer & RNA #splicing) - my own views only
Hola! We are the CRG, a research institute exploring the frontiers of biology in Barcelona. Som un centre CERCA. www.crg.eu


















