
Pretty neat to see our paper - led by @kr-maynard.bsky.social @lcolladotor.bsky.social on the 2021 list of most cited neuroscience papers 🧠🔬🧪
www.thetransmitter.org/publishing/w...
@lmweber.bsky.social
Assistant Professor of Biostatistics, Boston University. Statistical genomics, genomic data science, open science, R programming https://lmweber.org/

Pretty neat to see our paper - led by @kr-maynard.bsky.social @lcolladotor.bsky.social on the 2021 list of most cited neuroscience papers 🧠🔬🧪
www.thetransmitter.org/publishing/w...
This effort started back in the early Visium days with our colleagues at the @lieberinstitute.bsky.social including @martinowk.bsky.social @kr-maynard.bsky.social @lcolladotor.bsky.social and others. Wonderful to have a complete release version out now. Thanks to everyone who contributed! 🧬🧠🎉
22.11.2025 15:22 — 👍 6 🔁 1 💬 0 📌 0
Link to preprint here: www.biorxiv.org/content/10.1...
22.11.2025 12:55 — 👍 3 🔁 1 💬 1 📌 0This was a huge effort by many contributors, in particular @helucro.bsky.social @estellayixingdong.bsky.social @rgottardo.bsky.social @markrobinsonca.bsky.social @stephaniehicks.bsky.social , and the @bioconductor.bsky.social core team and community. Thank you everyone! 🎉
22.11.2025 12:38 — 👍 5 🔁 1 💬 1 📌 0This contains extensive code examples, workflows, datasets, and discussion on data analysis workflows for spatial transcriptomics data using Bioconductor in R. Please check it out - feedback and suggestions welcome! bioconductor.org/books/OSTA/
22.11.2025 12:38 — 👍 1 🔁 1 💬 1 📌 0We are excited to share our online book and preprint on “Orchestrating Spatial Transcriptomics Analysis with Bioconductor”!
22.11.2025 12:38 — 👍 18 🔁 12 💬 1 📌 0I am very excited to share this book! It has been so much to collaborate on the project. We hope you find it useful!
21.11.2025 22:11 — 👍 22 🔁 7 💬 0 📌 0Orchestrating Spatial Transcriptomics Analysis with Bioconductor https://www.biorxiv.org/content/10.1101/2025.11.20.688607v1
21.11.2025 14:46 — 👍 13 🔁 10 💬 0 📌 1

Orchestrating Spatial Transcriptomics Analysis with Bioconductor
(including interoperability with Python)
Paper: www.biorxiv.org/content/10.1...
Book: bioconductor.org/books/OSTA/
#Rstats
Whether you're new to bioinformatics or a seasoned pro, OSTA covers everything that you need to know about spatial transcriptomics data analysis spanning both sequencing and imaging-based technologies within the Bioconductor framework 👨💻🧬
Congrats to all the authors on this huge effort!
Honored to have gotten to be a part of this! I vividly remember the very early days of spatial transcriptomics when we were all scratching our heads over what to do with all this new data! It's come a long way - congrats @helucro.bsky.social @lmweber.bsky.social and everyone who contributed to this!
21.11.2025 18:44 — 👍 14 🔁 3 💬 0 📌 0Many people hours, calls and messages later: OSTA is now “in (pre)print”, though the real thing lives at bioconductor.org/books/OSTA.
Check it out, get in touch. We welcome any feedback, suggestions, wishes (& contributions).
It’s been a joy working with you @estellayixingdong.bsky.social!
so excited to share that our preprint characterizing the dorsal anterior cingulate cortex in post-mortem human brain from @lieberinstitute.bsky.social is out! we integrated matched SRT and snRNA-seq data to investigate agranularity and von Economo neurons
24.07.2025 01:53 — 👍 13 🔁 3 💬 1 📌 3
Winter in Western Australia ☀️
29.06.2025 12:38 — 👍 4 🔁 0 💬 0 📌 0
Going from methods dev to full on cancer bio has been tough. All the more excited to see this out at last—
www.biorxiv.org/content/10.1...
@pascual-reguant.bsky.social
@brukerspatial.bsky.social
@hoheyn.bsky.social

Update: We greatly revised our paper and renamed it “Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta”.
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561
Excited to launch our AlphaGenome API goo.gle/3ZPUeFX along with the preprint goo.gle/45AkUyc describing and evaluating our latest DNA sequence model powering the API. Looking forward to seeing how scientists use it! @googledeepmind
25.06.2025 14:29 — 👍 220 🔁 82 💬 5 📌 10Very excited to see this important work out led by the talented @mictott.bsky.social @boyiguo.bsky.social @jhubiostat.bsky.social! 🧪🧬🖥️
06.06.2025 17:34 — 👍 22 🔁 5 💬 1 📌 0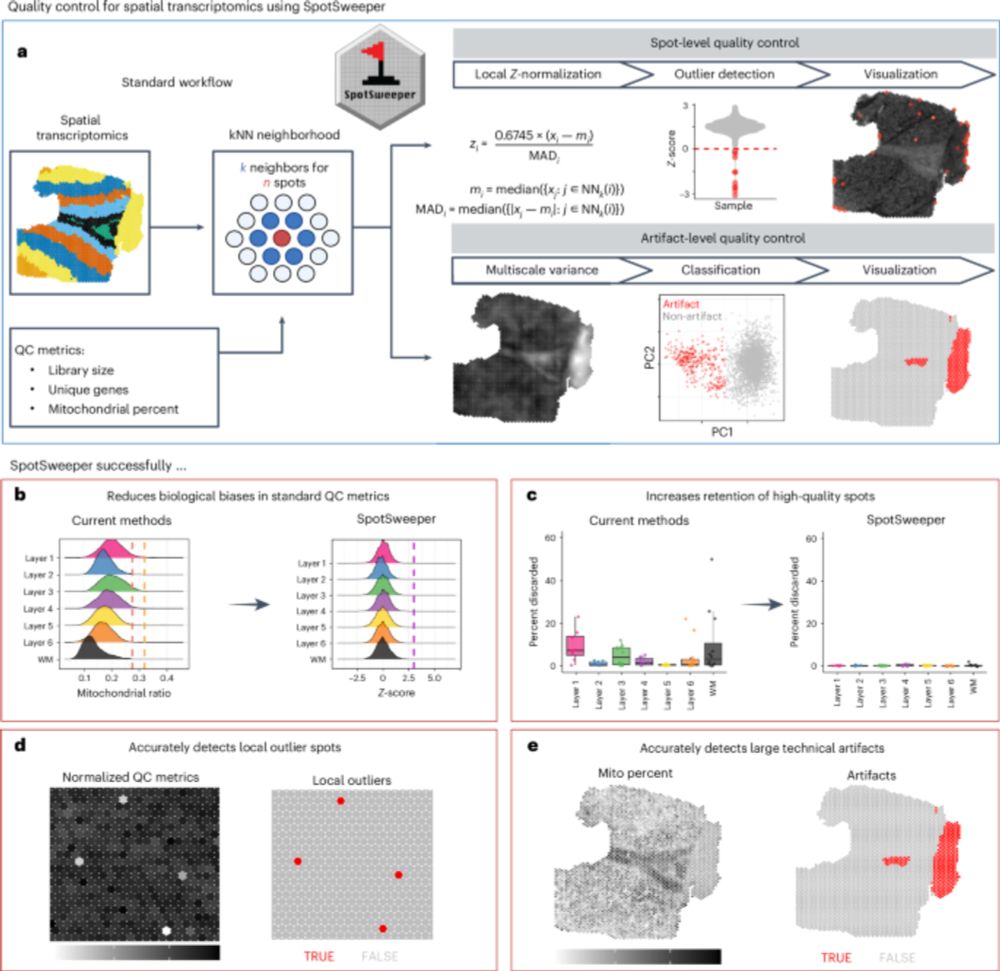
🚨 New paper published in @natmethods.nature.com!
We introduce SpotSweeper, the first spatially-aware QC methods for spatial transcriptomics.
📰 Paper : nature.com/articles/s41...
💻 Code: github.com/MicTott/Spot...
📈 Website: mictott.github.io/SpotSweeper/
🧵👇
my work with @stephaniehicks.bsky.social and @boyiguo.bsky.social has been published in Biostatistics! tldr: the mean-variance bias exists in SRT data & impacts SVG detection. we propose #spoon, available on @bioconductor.bsky.social, to address this! pmc.ncbi.nlm.nih.gov/articles/PMC... 🤩
17.06.2025 00:17 — 👍 7 🔁 3 💬 2 📌 0Congratulations!
24.06.2025 01:11 — 👍 1 🔁 0 💬 1 📌 0Congratulations!! 🎉
23.06.2025 02:56 — 👍 1 🔁 0 💬 0 📌 0Feels like a social media tipping point might have happened 🐦🔵
13.11.2024 21:36 — 👍 8 🔁 0 💬 1 📌 0

This morning I'm teaching an intro to single cell data science!
Feeling very appreciative to all the amazing developers in the #Bioconductor and open science #rstats communities! #Genomics 🖥️ 🧬 #stats 👩🔬
🔗 stephaniehicks.com/singlecellde...

One last visit to Washington DC before I leave for Boston 🇺🇸
04.09.2023 23:03 — 👍 2 🔁 0 💬 0 📌 0
Hello world 🙂🌤️
01.09.2023 19:09 — 👍 11 🔁 0 💬 2 📌 0