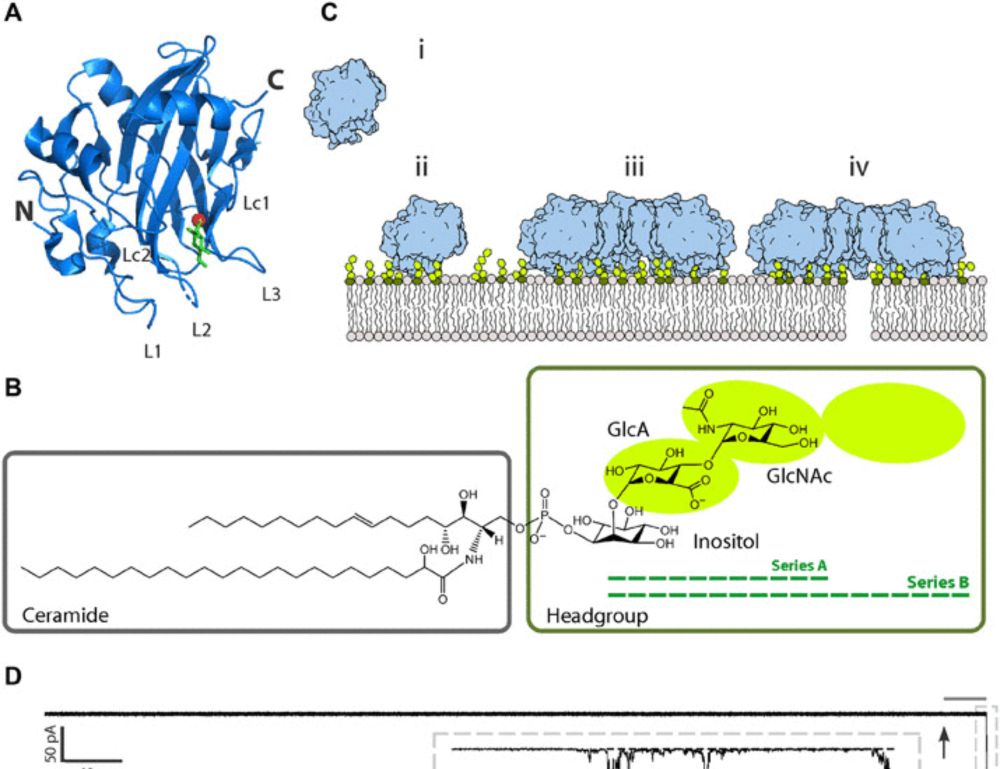
Almost at the end of the exciting @cecamevents.bsky.social conference 'RNA Modelling across scales' at SISSA, Trieste, Italy! 🎉 Fantastic speakers and great science. Organized with @bussigio.bsky.social @marcodevivo.bsky.social #RNA
21.05.2025 19:22 — 👍 19 🔁 8 💬 0 📌 3
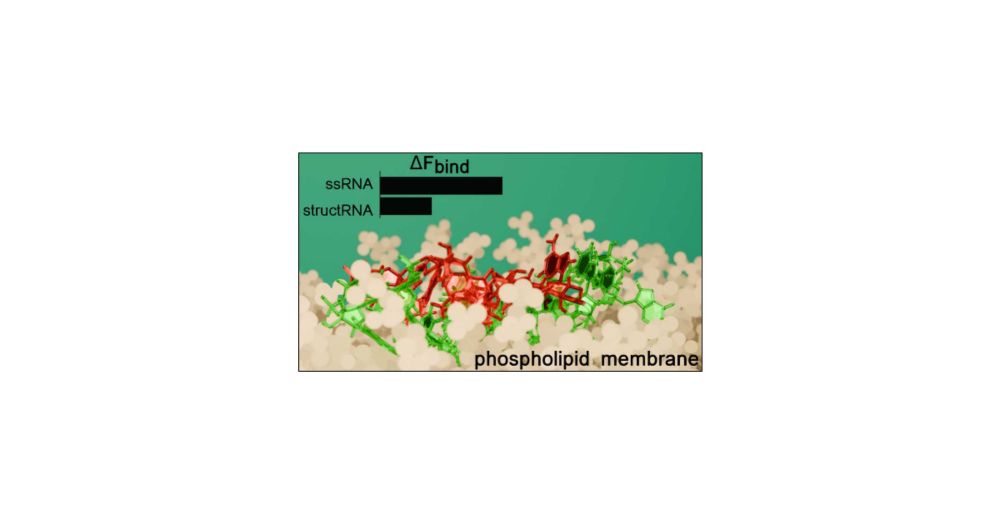
Our integrative approach using #MD and #cryoEM data to construct structural ensembles of #RNA just published on @natcomms.bsky.social doi.org/10.1038/s414... Lead by Elisa Posani, with @magistratolab.bsky.social @bonomimax.bsky.social @pjanos.bsky.social @navtejtoor.bsky.social and Daniel Haack 🎉
16.05.2025 09:16 — 👍 26 🔁 8 💬 0 📌 0

Check out the speakers!
@rommieamaro.bsky.social @jessicaandreani.bsky.social @gauravbhardwaj.bsky.social @bonomimax.bsky.social @rocovino.bsky.social @arneelof.bsky.social Sergei Grudinin @magistratolab.bsky.social Christine Orengo @joanampereira.bsky.social Maya Topf, Sameer Velankar, Basile Wicky
09.05.2025 10:37 — 👍 9 🔁 5 💬 0 📌 0
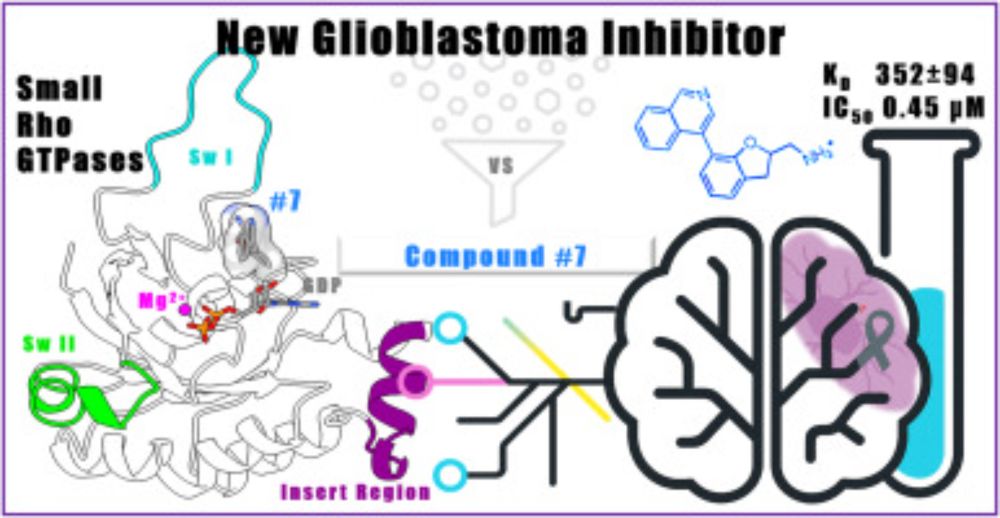
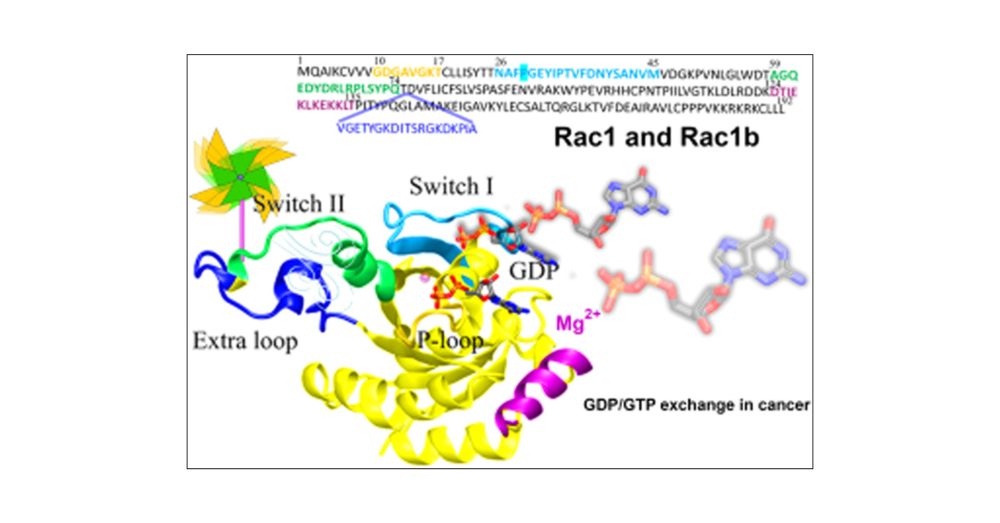
Identification of a new Small Rho GTPase Inhibitor Effective in Glioblastoma Human Cells
Glioblastoma (GBM) is the most common and lethal primary brain tumour. The prognosis for GBM patients remains poor due to rapid tumour recurrence and …
Just published our new paper on Identification of a new Small Rho GTPase Inhibitor Effective in Glioblastoma Human Cells. Check it out! www.sciencedirect.com/science/arti.... Excellent work done by
@angelaparis3.bsky.social , in collaboration with University of Trieste, University of Udine, CNR-IC
05.05.2025 11:54 — 👍 7 🔁 2 💬 0 📌 0
📢 Our article calling for a #FAIR database for #MolecularDynamics simulation data has now been peer-reviewed and published in @naturemethods.bsky.social
📖 Read it here: rdcu.be/ef6YX
📝 Support the statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #collaboration
04.04.2025 08:09 — 👍 37 🔁 21 💬 0 📌 3
We are sill accepting applications for poster presentations at the CECAM conference 'RNA Modelling Across Scales". Join us in the beautiful Trieste ☀️🏖️🏰! Apply now: www.cecam.org/workshop-det... Co-organized with @bussigio.bsky.social @marcodevivo.bsky.social
27.03.2025 19:04 — 👍 10 🔁 5 💬 0 📌 0
CECAM - RNA modelling across scales
Interested in #RNA modelling across multiple scales? Join us for the @cecamevents.bsky.social meeting at SISSA, Trieste, May 19-22! www.cecam.org/workshop-det... We still have slots for contributed talks. Deadline March 23. Co-organized with @magistratolab.bsky.social and @marcodevivo.bsky.social
20.02.2025 19:22 — 👍 18 🔁 11 💬 0 📌 1

News - EuChemS
WINNER ANNOUNCEMENT! “EuChemS ChemBioLife Young Investigator Award” 2024-2025 edition We are proud to announce the winner of the EuChemS ChemBioLife Young Investigator Award of the EuChemS Division...
Very proud to announce that a member of our group, Dr. Jana Aupič, has been awarded the prestigious "EuChemS ChemBioLife Young Investigator" prize! 🏅 This honors her hard work and exceptional contribution to chemical biology. Congratulations, Jana! 🌟✨ .
www.euchems.eu/divisions/ch...
04.02.2025 16:03 — 👍 10 🔁 5 💬 0 📌 0

✨Great presentation of our work by @angelaparis3.bsky.social at Physics Day in Trieste!👏
13.12.2024 15:17 — 👍 6 🔁 1 💬 0 📌 0
Associate Professor (University of Valencia 🇪🇸)
Computational chemistry and photocatalysis
Computational biophysicist @ Max Planck Institute for Polymer Research, Mainz.
Cheminformatician/computational-biochemist working on drug discovery (señor postdoc, CMD-PX / OPIG, University of Oxford)
⌬ ⚗️💻
Author of michelanglo.sgc.ox.ac.uk and Fragmenstein
Franco Lab @UCLA Exploring how to program dynamic behaviors in biological self-assembly 💡✨ We’re harnessing DNA and RNA to build smart, adaptive materials 🔬🧬 #Biomaterials #DNA #RNA
Computational Structural Biology, The Hebrew University of Jerusalem
Protein Disorder in Transcription Lab of Sina Wittmann at @imbmainz.bsky.social
Molecular Modeling & Drug Design. The lab of Stefano Forli at Dept. of Integrative Structural & Computational Biology at @scripps.edu
Home of the AutoDock
https://forlilab.org/
Computational structural biology, drug discovery, antibodies, biophysics, AI/ML, and stuff (mostly about proteins)
Lorenzo Di Michele's research group at @cebcambridge.bsky.social University of Cambridge (and also a bit at Imperial College). Working on DNA/RNA nanotechnology and Synthetic Cells
We study RNAs and RNA-binding proteins with a strong focus on neurodegeneration. Our research combines theoretical and experimental approaches to investigate liquid–liquid phase separation and liquid–solid phase transitions of protein and RNA molecules.
ICREA Research Professor at IRB Barcelona. Working on computational cancer genomics. Leading @bbglab.bsky.social
http://bbglab.irbbarcelona.org
Structural biology and pharmacology, with a focus on GPCRs and membrane transporters. 💊 🧪
https://www.gati-lab.com
https://scholar.google.com/citations?user=5t80YUAAAAAJ&hl=en
GōMartini 3 developer. 🇵🇪 Dad. Fascinated by how pathogens interact with cells.
🌐 http://pomalab.ippt.pan.pl/web/
Multiscale molecular dynamics, biological membranes interactions, CNRS researcher, group leader at @cbitoulouse.bsky.social
A group of computational chemists at UCLA. We study catalysts, materials, and complex (bio)molecular systems (and make them better!).
Student-run account.
https://alexandrova.chem.ucla.edu/
nucleic acids, polymerases, helicases, and mitochondria at Tufts University
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰
Achira | http://achira.ai
Research laboratory | http://choderalab.org
Antiviral drug discovery for pandemics | http://asapdiscovery.org
OpenADMET | http://openadmet.org
Employer-mandated disclaimer: http://choderalab.org/disclaimer
Pronouns: he/him
Prof of Computational Microbiology. Oxford Biochemistry & St Anne’s College. Bacterial Cell Envelopes. LFC fan 🌈
Chemistry professor at CMU. Connecting chemical sciences with AI #MachineLearning and automated experimentation. #tarheels fan. Care: #design, #photography #Ukraine #cats🐈 Rants are mine