And as always, I'm happy to answer any questions or receive any feedback on this work!
02.09.2025 18:32 — 👍 1 🔁 0 💬 0 📌 0
I hope Sam (who is a math/economics major, and had never taken a genetics class before this project) learned as much about polyploids as I learned from him about math. Thanks also to @ryangutenkunst.bsky.social and @barkerms.bsky.social for overseeing this project as well
02.09.2025 18:32 — 👍 4 🔁 0 💬 1 📌 0
Overall, I'm super excited about this paper, as it is one of the first pieces of theory based on tetraploids with disomic inheritance, without treating the subgenomes independently. I think there is still lots to learn on this front, and hope that other will help push this field forward
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
Here, we can directly measure additive genetic variance and the change in allele freq and pop fitness. When the population is in the bistability regime, Fisher's Fundamental Theory of Natural Selection does not hold, and there are subtle differences between disomic and polysomic inheritance patterns
02.09.2025 18:32 — 👍 3 🔁 0 💬 1 📌 0
Finally, we wanted to see if there was a difference to how polyploid populations might approach these equilibrium points based on segregation patterns. For this, we developed a forward-in-time 1 locus simulation for disomic and polysomic inheritance.
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
The ability for a population to remove this is totally dependent on whether the initial allele frequency is higher or lower than the unstable equil point. Also, because this model assumes infinite population sizes, there is no influence of drift in this model.
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
This is a classic example of a saddle node bifurcation in nonlinear dynamics. Meaning, a population may be completely unable to purge a deleterious mutation, despite it being dominant and at high/intermediate frequency, depending on the initial allele frequency. Exactly the opposite of masking.
02.09.2025 18:32 — 👍 4 🔁 0 💬 1 📌 0

When the deleterious mutation rate is higher than the back-mutation rate (in this case, 40x greater), then we see a totally different dynamic for dominant muts - for some selection coefficients, there is no single equilibrium point - there are three points! Two are stable points, one is unstable
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
This 'bump' appears to coincide with the point at which the distribution of genotype frequencies is the farthest from Hardy-Weinberg Equilibrium - suggesting another avenue in which the allele frequency is insufficient to describe population dynamics in polyploid populations
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
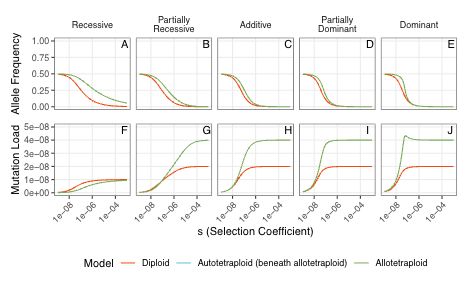
First, we found that our results are quite sensitive to mutation rates. If the deleterious mutation rate = back mutation rate, then this is the plot of the equilibrium allele frequency and load of the populations. Nothing surprising here, except there is a 'bump' in the load for the dominant case
02.09.2025 18:32 — 👍 3 🔁 0 💬 1 📌 0

A mathematical equation that represents the first order partial differential equation for the change in gamete frequencies for a tetraploid with polysomic inheritance. The expression is long, and is in a tiny text, to highight that it is impossible to intuit anything from this expression alone.
These PDEs are not easy to analytically understand, and are surprisingly much much more complex than the diploid ODEs (it's not just a factor of two difference!). This is just one of the entries of the Jacobian for a tetraploid with polysomic inheritance - don't stare at this for too long, I beg you
02.09.2025 18:32 — 👍 4 🔁 0 💬 1 📌 0
In order to probe this question, Sam had to approach it using a nonlinear dynamics framework. Although mut-sel equilibrium is easy in diploids (and can be analyzed using a simple ODE), for polyploids, Sam had to develop a framework to numerically estimate equilbria points with a series of PDEs
02.09.2025 18:32 — 👍 3 🔁 0 💬 1 📌 0
And, the only theory work that has been done for any tetraploid is for very idealistic and convenient dominance relationships (completely recessive, perfectly additive) that make the math simpler. We wanted to look at arbitrary dominance relationships, to see if any interesting patterns arose
02.09.2025 18:32 — 👍 2 🔁 0 💬 1 📌 0
Although mut-sel equilibria has been estimated for tetraploids with polysomic inheritance, very little theory has been developed for tetraploids with disomic inheritance. We wanted to see if segregation differences changed these mut-sel equilibrium points, or how populations approach these points
02.09.2025 18:32 — 👍 3 🔁 0 💬 1 📌 0
That's true, with a strong enough drawl, there's no confusing the two. There is a flow cytometer here that is part of the center's shared equipment, it's currently not being used. So it'll migrate down the hallway into my space as soon as I get it ready
21.08.2025 13:10 — 👍 1 🔁 0 💬 0 📌 0
Annie Mae might evolve into Anime by accident? Or on purpose, your choice
20.08.2025 23:13 — 👍 3 🔁 0 💬 1 📌 0
Oh I'm stealing this for the flow cytometer my lab is inheriting!
20.08.2025 23:12 — 👍 2 🔁 0 💬 0 📌 0
Overall, I'm excited to see future broader sampling of high-quality genomes refine the story we've shown here, arguably one of the most complicated polyploid histories in plants. And, there's a ton to be learned about how this ancient polyploid history influences modern biological processes, too!
13.08.2025 20:31 — 👍 6 🔁 0 💬 1 📌 0
We found a bunch of traits are associated with this ancient polyploid history, and especially with the rate at which chromosome number (and gene content) is reduced, suggesting that the 'recovery' that lineages much endure from this massive ploidy increase also leads to, e.g., higher speciation rate
13.08.2025 20:31 — 👍 2 🔁 0 💬 1 📌 0
And yet, one of the genomes assembled for this paper (Reevisia pubescens) has a totally different polyploid history, even though it is nestled in the middle of this polyploid rich family (it was in the same subfamily as Durio! This is also the reason we suggest a new subfamily designation for Durio)
13.08.2025 20:31 — 👍 1 🔁 0 💬 1 📌 0
In this new paper, which has a genome from every subfamily, we discovered that the polyploid history looks to be one giant mess - All three subgenomes of the hexaploid Durio are found in the decaploid cotton, and 2 of the 3 subgenomes in Durio can be found in subfamilies that only have a duplication
13.08.2025 20:31 — 👍 3 🔁 0 💬 1 📌 0
When the Durio zibethinus genome came out in 2017, it showed evidence of a whole genome triplication. So not the exact same history as cotton, but we couldn't rule out if that hexaploidy was a stepping stone on the same path that cotton took on its way to become decaploid (10N) or dodecaploid (12N)
13.08.2025 20:31 — 👍 1 🔁 0 💬 1 📌 0
When the first diploid cotton genome was published in 2013, there was evidence of an ancient 5- or 6-fold multiplication that wasn't shared by the chocolate genome (one of the early-diverging lineages to the rest of the family). But it wasn't known whether this was a singular or multiple events.
13.08.2025 20:31 — 👍 2 🔁 0 💬 1 📌 0
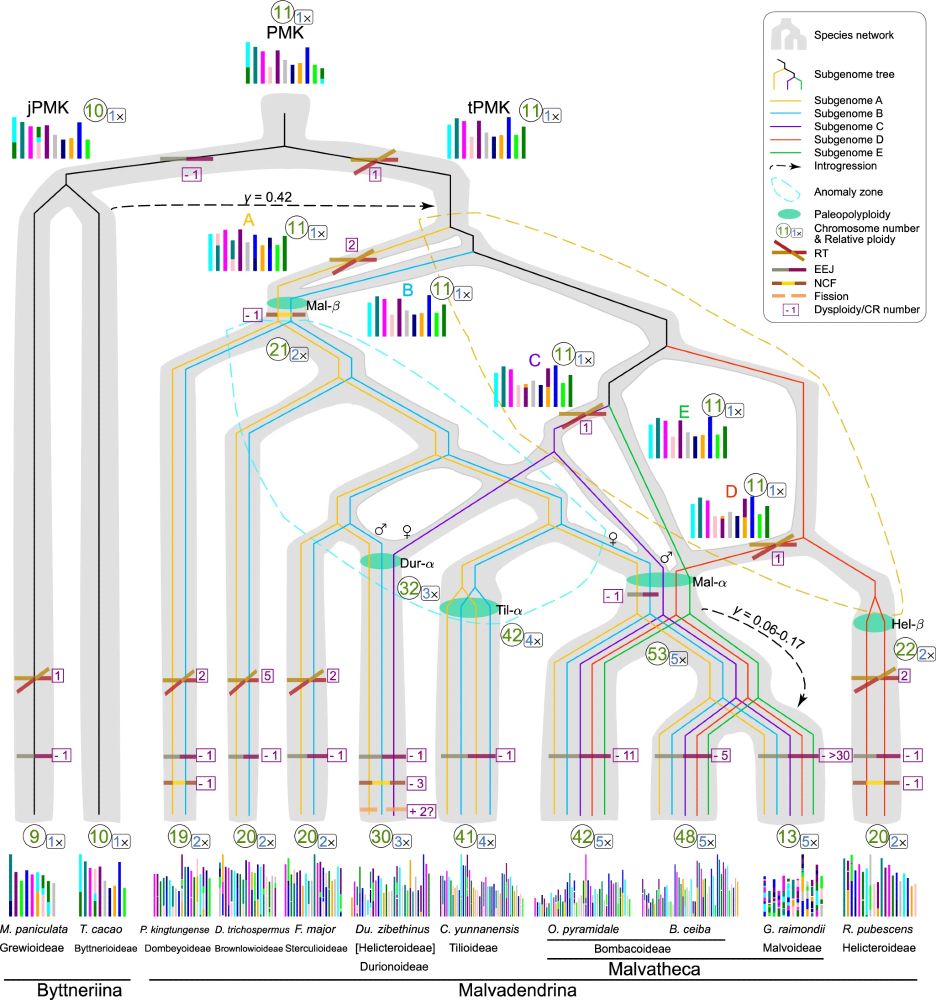
New Paper! In 2019, my first dissertation chapter revealed complicated polyploidies in the cotton family (Malvaceae) but we lacked the tools and genomes to truly understand it. Now with better genomes and improved methods, it's much more complicated than we thought. www.nature.com/articles/s41...
13.08.2025 20:31 — 👍 71 🔁 20 💬 3 📌 1
Oh this is really fun!
05.08.2025 21:33 — 👍 1 🔁 0 💬 0 📌 0

Logo for the Genomic History Inference Strategies Tournament.
If you're new to demographic history inference from population genomics, try this webapp I created to illustrate how dadi fits bottleneck models to site frequency spectra: ryangutenkunst-dadi-two-epoch.hf.space . It even outputs files for submitting to the GHIST competition! ghi.st
05.08.2025 18:48 — 👍 39 🔁 21 💬 1 📌 2
The Conover Lab is open!
I am so excited to join a group of talented, caring, and creative folks at the @danforthcenter.bsky.social, all in service of improving the human condition through plant science.
01.07.2025 15:48 — 👍 46 🔁 8 💬 1 📌 0
All you need is read counts for positions you want to call SNPs for. We used GATK for this, but any program that can give allele counts for read mapping works. Feel free to reach out and I'm happy to help debug!
12.06.2025 22:46 — 👍 4 🔁 0 💬 0 📌 0
GitHub - dcgerard/updog at rm
Flexible Genotyping of Polyploids using Next Generation Sequencing Data - GitHub - dcgerard/updog at rm
We're writing up the paper now, but all of the code is publicly available on the rm branch of the github repo, and works for any even-ploidy population. github.com/dcgerard/upd...
12.06.2025 22:46 — 👍 3 🔁 0 💬 1 📌 0
Montana State Plant Sciences & MONT herbarium
NSF Postdoc at UC Santa Cruz, studying population and conservation genomics, and taking photos of my cats. she/her
Curator at the New York Botanical Garden. Plants, rivers, phylogenetics, extreme botany and arepas
https://anamariabedoya.weebly.com
Evolutionary biologist, computational biologist, statistician. I like to develop mathematical models of evolutionary process and see how they fit to data. I also like cities where building apartments is legal.
PhD student at Michigan State’s Kellogg Biological Station researching spotted turtle phylogeography and the evolution of temperature-dependent sex determination
Computational biology and evolutionary genetics. PhD in Biology at the University of Oregon with Andrew Kern and Peter Ralph. From 🇧🇷. https://m-rodrigues.me
Botanist. University of Georgia. Plant evolutionary biology, phylogenomics, comparative genomics.
Evolutionary Biology, Phylogeography, Ichthyology, Conservation Genetics, Community Ecology, Friend of Rough Fish
Postdoc @ucsf-bchsi.bsky.social with Tony Capra. PhD Berkeley with Rasmus Nielsen. ARGs, computational medicine, stat. computing, ancient DNA. https://github.com/avaughn271
Postdoc at the San Diego Zoo Wildlife Alliance interested in population genetics & conservation genomics https://chriskyriazis.weebly.com/
assistant prof at University of Oregon. interested in pop gen, stat gen, human complex traits. also ELSI, metascience, ethics education, etc...
she/they. 🌈
roshnipatel.github.io
Evolutionary and community ecologist studying figs and the things that live in them. Assistant Professor at University of Missouri. Also into metal.
Assistant Professor, University of Arizona School of Natural Resources & the Environment | PopGen, Phylogenomics, Conservation Genomics | she/her
Postdoc @uwgenome 🐶
PhD Genetics @uarizona 🐱
BA Chemistry @brynmawrcollege 🦉
Population Genomics, Machine Learning, Cancer Evolution 🧬💻
📍Seattle, WA & Vancouver, BC
https://www.linkedin.com/in/linhnhtran/
E. Desmond Lee Professor in Botanical Studies #UMSL #MOBOT | #PopulationEcology #MacrosystemsBiology #ClimateChange #MastSeeding #ForestEcology #UrbanEcology | #FirstGen she/her 🇨🇦 in St. Louis
lamontagnelab.weebly.com
Assistant professor of evolutionary genomics at KU. Has a toddler 🙂🙃🙂🙃🇰🇷🇨🇦🇺🇲
https://jychoilab.github.io/
Passionately researching to understand the transport processes and genes required for proper chloroplast function and photosynthesis.
Treasuring exceptions at IJPB (INRAE Versailles). Thirsty for genetics, chromosome biology and evolution.
Ecological differentiation, genetic elements, and speciation in dune-endemic sunflowers🌻
PhD Student, Ostevik Lab 🌱
EEOB Department at UC Riverside
A free pre-print and post-print service for ecology, evolution, and conservation. Run by @sortee.bsky.social.
Server: https://ecoevorxiv.org
Instructions & FAQ: https://sortee.github.io/ecoevorxiv/






