Very much looking forward to our 10th (anniversary!) Microbial Genomics Training Course provided by #BiGi @denbi.bsky.social in Gießen.
3 days full of free hands-on microbial genomics training - from QC, assembly and annotation to genome submissions and comparative genomics.
27.02.2026 15:42 —
👍 3
🔁 3
💬 0
📌 0


🔬✨ Tool of the week: #Metagenomic data analysis made easy!
🚀 The Metagenomics-Toolkit offers a complete workflow: from raw Illumina or Nanopore reads 📈 to taxonomic 🦠 and functional 🧬 insights.
📊 Reproducible, publish-ready results.
Ready to boost your research? 🌍📝
www.denbi.de/services-acc...
24.02.2026 07:36 —
👍 4
🔁 2
💬 0
📌 0

AlphaFold Database welcomes community datasets
Latest AlphaFold Database update adds high-value datasets for microbial and viral proteins, generated by specialist communities
Delighted to see over 17 million new protein structure predictions from novel proteins in AllTheBacteria are now integrated into the AlphaFold Database at @ebi.embl.org !
Huge work from @gbouras13.bsky.social @oschwengers.bsky.social and friends to generate these.
www.ebi.ac.uk/about/news/u...
17.02.2026 13:52 —
👍 97
🔁 26
💬 1
📌 2

Release v1.12 - Just do it, but don't crash · oschwengers/bakta
This is the twelfth minor release (v1.12) providing more than 10 minor improvements and many bug fixes improving runtime stability, IO compatibility, and last but not least user experience.
Compati...
🦠🧬🖥️ Bakta v1.12.0 is out
with tons of tiny improvements and bug fixes, too many to list all:
- partial genes on linear seqs
- improved errror handlings & runtimes
- support Python 3.12 & 3.13
- ...
A huge shout out and thank you to all bug reporters and contributors!
github.com/oschwengers/...
02.02.2026 08:05 —
👍 9
🔁 7
💬 0
📌 1
Honestly, as a micro binfie, I couldn't feel more honored than to be entrusted with this! 😃
Prokka has served so many so well, and deeply inspired me how to write Bakta.
I'm thankful to be part of this open science com, and will do my best to meet these high quality standards.
Thanks Torsten! 🦠💻
16.12.2025 11:50 —
👍 28
🔁 3
💬 0
📌 1

We are de.NBI & ELIXIR-DE, came together for our All Hands 2025! #deNBI2025 #ELIXIRDE #AllHands
24.11.2025 09:30 —
👍 6
🔁 4
💬 0
📌 0
We would also like to deeply thank @denbi.bsky.social
for the lasting support and funding enabling constant maintenance and development!
06.10.2025 08:11 —
👍 1
🔁 1
💬 1
📌 0

Unbelievable, Bakta reached its 1,000th citation!
A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!
06.10.2025 08:11 —
👍 46
🔁 10
💬 1
📌 0
Isolates or MAGs, we're looking for the worst-annotated genomes, i.e. high fraction of CDS w/o any functional annotation. Any public/shareable bacterial genome is highly welcome!
Please help us to further improve the annotation of bacterial genomes.
Please RT & share
(2/2)
06.10.2025 07:27 —
👍 4
🔁 2
💬 0
📌 0
Dear community, Bakta needs your help!
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
06.10.2025 07:27 —
👍 11
🔁 18
💬 1
📌 0
Definitely the best to put next. We'll soon start some internal discussion of how/when to integrate that. It's been a pleasure to meet you in person Zam!
22.09.2025 11:13 —
👍 1
🔁 0
💬 0
📌 0
Thanks so much Migle! It's been a pleasure to meet you at IMMEM and I'm glad you find BakRep useful!
22.09.2025 11:11 —
👍 1
🔁 0
💬 0
📌 0
9th Microbial Genomics training course
The 'German Network for Bioinformatics Infrastructure – de.NBI' is a national, academic and non-profit infrastructure supported by the Federal Ministry of Education and Research providing bioinformati...
Our 9th @denbi.bsky.social "Microbial Genomics training course" is now open for registration!
3 sessions full of microbial bioinformatics at JLU Giessen:
I: QC & QA, assembly
II: regional & functional annotation
III: comparative genomics
Info & registration: www.denbi.de/training-cou...
08.05.2025 13:08 —
👍 3
🔁 3
💬 0
📌 0
100% agreed - exactly that was also our line of thinking.
28.04.2025 09:54 —
👍 1
🔁 0
💬 0
📌 0
Thanks @kblin.bsky.social for pointing out. Of course, it is not a proper encryption, however hard to guess.
So, please use a proper encryption for sharing - if required.
28.04.2025 09:26 —
👍 0
🔁 0
💬 1
📌 0
Bakta
Even without registration, results can easily and safely be shared with others using encrypted URLs.
Of course, your data is handled with care automatically discarded after 30 days.
Just give it a try: bakta.computational.bio
(5/5)
28.04.2025 08:47 —
👍 1
🔁 0
💬 0
📌 1
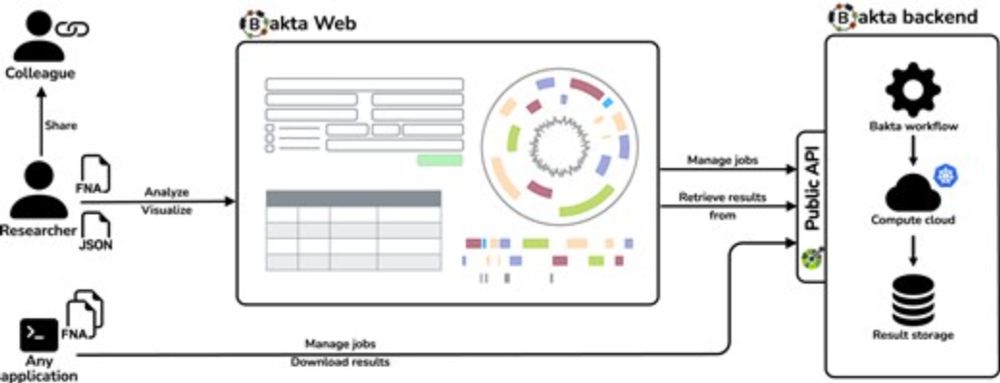
Results are presented via a cross-referenced data table, a linear genome browser and circular genome plots - just choose your viz!
Of course, all results are also available for download in various standard file formats: GFF3, GenBank, EMBL, FASTA, TSV, TXT, JSON…
(4/5)
28.04.2025 08:47 —
👍 1
🔁 0
💬 1
📌 0
All executions are conducted in a scalable Kubernetes backend hosted within the @denbiOffice cloud.
By this, Bakta Web dynamically scales out to meet temporal peak demands.
(3/5)
28.04.2025 08:47 —
👍 1
🔁 0
💬 1
📌 0
Bakta Web is our web application accompanying Bakta’s CLI version.
It provides interactive widgets supporting users with both, the provision of input data and sample/sequence metadata, and the interpretation of annotation results.
(2/5)
28.04.2025 08:47 —
👍 1
🔁 0
💬 1
📌 0
Bakta Web – rapid and standardized genome annotation on scalable infrastructures
Abstract. The Bakta command line application is widely used and one of the most established tools for bacterial genome annotation. It balances comprehensiv
We happily present: “Bakta Web – rapid and standardized genome annotation on scalable infrastructures” @OxUniPress NAR’s Web Server issue
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
28.04.2025 08:47 —
👍 42
🔁 28
💬 1
📌 0

Interested in bacterial genomes?
Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25
25.03.2025 09:45 —
👍 18
🔁 9
💬 2
📌 0
Of course, the DB light version was also updated, however DB sizes have barely changed.
(6/6)
06.03.2025 08:59 —
👍 0
🔁 0
💬 0
📌 0
Due to this massive increase in known protein sequences, DB size has grown from 72GB to 83GB (deflated).
To mitigate negative download effects for our users, we therefore switched tarball compressions from gzip to xz.
Thus, compressed DB size actually decreased from 37GB to 30GB!
(4/6)
06.03.2025 08:59 —
👍 0
🔁 0
💬 1
📌 0
To further improve annotation quality, external annotation sources have also been updated:
- COG: 2020 -> 2024
- Pfam: 36 -> 37.2
- RefSeq: r220 -> r228
- Rfam: 14.10 -> 15
- UniProtKB: 2023_05 -> 2025_01
Also, NCBI AMRFinderPlus and VFDB have been updated to the most recent versions.
(3/6)
06.03.2025 08:59 —
👍 1
🔁 0
💬 1
📌 0
This is a massive increase in known protein sequences - which is great!
Numbers of IPS and PSC have grown by 22% and 13%.
(2/6)
06.03.2025 08:59 —
👍 0
🔁 0
💬 1
📌 0
Validate User
Our paper on bacterial #Resistance prediction with #Genomics and #MachineLearning reached a milestone with its 120th citation! 🎉 @oschwengers.bsky.social
academic.oup.com/bioinformati...
05.03.2025 19:10 —
👍 4
🔁 1
💬 0
📌 0