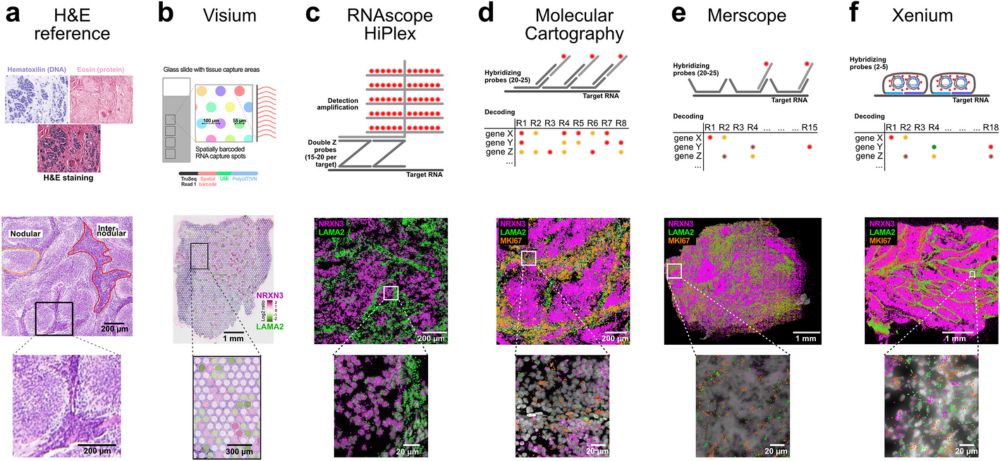
8/ 🤘 For those about to rock (around the Christmas tree), we salute you 🎸🎄: Isabelle Seufert & our team from @dkfz.bsky.social & BioQuant & @akispapantonis.bsky.social, Philipp Mallm, @s-anders.bsky.social & Petros Kolovos labs with support from @spp2202.bsky.social & @mspacealliance.bsky.social.
24.12.2025 22:54 — 👍 3 🔁 1 💬 1 📌 0

7/ 🌟 The broad applicability of our RWireX framework was shown in different cellular systems. In human blood cells, cell-type-specific regulatory programs could be distinguished, e.g., the DCs at the CD22 and the TBX21 loci had highest activity in B cells and mature CD8+ T cells, respectively.
24.12.2025 22:52 — 👍 1 🔁 0 💬 1 📌 0

6/ 🎯 Our AC/DC framework reveals how cells achieve precise gene control. TRGs with TF promoter binding show fastest induction. DCs enable coordinated burst-size increases, while ACs allow fine-tuned gene-specific regulation—working together to support complex programs.
24.12.2025 22:51 — 👍 3 🔁 1 💬 2 📌 0

5/ ⛰️ DCs are larger contiguous regions of simultaneously accessible chromatin sites. These domains exhibit locally increased transcription factor binding activity and may represent the genomic footprints of TF hubs.
24.12.2025 22:49 — 👍 2 🔁 0 💬 1 📌 0

4/ 🔗 ACs denote interactions between separated regulatory sites at ATAC peaks. We observed differences between rare and frequently occurring ACs, suggesting distinct roles of stochastic vs. architectural links.
24.12.2025 22:47 — 👍 2 🔁 0 💬 1 📌 0

3/ 💻 Using our new RWireX software (github.com/RippeLab/RWi...), we distinguish two regulatory chromatin modules: ACs (autonomous links of co-accessibility between separated sites of 1 kb size) and DCs (domains of contiguous co-accessibility of ~200 kb) that both can co-exist.
24.12.2025 22:46 — 👍 2 🔁 0 💬 1 📌 0

2/ 🔍 We investigated the proinflammatory response to TNF in primary human cells using #scRNA and #scATAC sequencing and fluorescence microscopy. We identified ~1,500 TNF-regulated genes (TRGs) that often clustered in the genome and exhibited induced co-expression.
24.12.2025 22:45 — 👍 3 🔁 0 💬 1 📌 0

1/ 🎄 We got our Christmas present today: "Two distinct chromatin modules regulate proinflammatory gene expression" is now published @natcellbio.nature.com doi.org/10.1038/s415.... Our study introduces a scATAC-seq-based framework for genome-wide analysis of gene regulation features.
24.12.2025 22:44 — 👍 77 🔁 24 💬 3 📌 2
Single-cell Open Lab - German Cancer Research Center
Happy to report that the Xenium system for spatial transcriptomics in the Single Cell Open Lab www.dkfz.de/en/single-ce... @dkfz.bsky.social was the first one worldwide to pass the mark of 100 runs according to @10xgenomics.bsky.social. And it keeps on running with another one, so stay tuned...
21.08.2025 16:12 — 👍 3 🔁 1 💬 0 📌 0
These features allow it to predict immunotherapy response and point to potential universal T cell based therapies. Great multi-group effort led by Mirco Friedrich together with @michael-platten.bsky.social, @raabms.bsky.social, @stefaneichmueller.bsky.social, and many more collaborators.
13.08.2025 13:00 — 👍 2 🔁 0 💬 0 📌 0

Glad that the preprint on identifying and characterizing rare tumor-reactive T cells in multiple myeloma & AML patients is out now doi.org/10.1101/2025.... Single-cell TCR and transcriptome profiling & immunopeptidomics revealed conserved programs and shared non-canonical antigens.
13.08.2025 12:58 — 👍 3 🔁 0 💬 1 📌 0

Liquid-liquid phase separation of HP1? Not in our mouse fibroblasts… This paper by the Fabian Erdel group offers a fresh perspective on how this can be reconciled with seemingly conflicting reports: The phase separation propensity of HP1 decreases from yeast → fly → mouse. doi.org/10.1038/s414...
11.07.2025 05:35 — 👍 32 🔁 13 💬 1 📌 0

3/ Great joint effort with Philipp Mallm and the Single Cell Open Lab @dkfz.bsky.social, led by Anne Rademacher, Alik Huseynov, and Michele Bortolomeazzi, with key contributions from our colleagues at KiTZ Heidelberg, supported by the @mspacealliance.bsky.social program of @hlsalliance.bsky.social.
20.06.2025 19:37 — 👍 2 🔁 0 💬 0 📌 0

2/ Using medulloblastoma cryosections, we analyzed sensitivity, specificity, and spatial signal patterns to distinguish background from rare-cell signals. We also show how reimaging slides improves cell segmentation—and enables additional transcript and protein readouts from the same section.
20.06.2025 19:34 — 👍 1 🔁 0 💬 1 📌 0
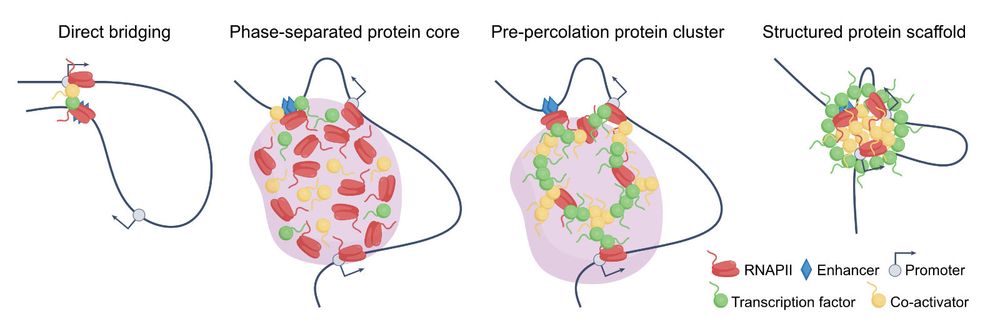
How do transcription compartments form — and does phase separation drive gene expression? I enjoyed discussing these questions with @akispapantonis.bsky.social a lot, and we put our thoughts together for @naturerevgenet.bsky.social, now out at rdcu.be/erP1u
19.06.2025 18:28 — 👍 53 🔁 14 💬 1 📌 1
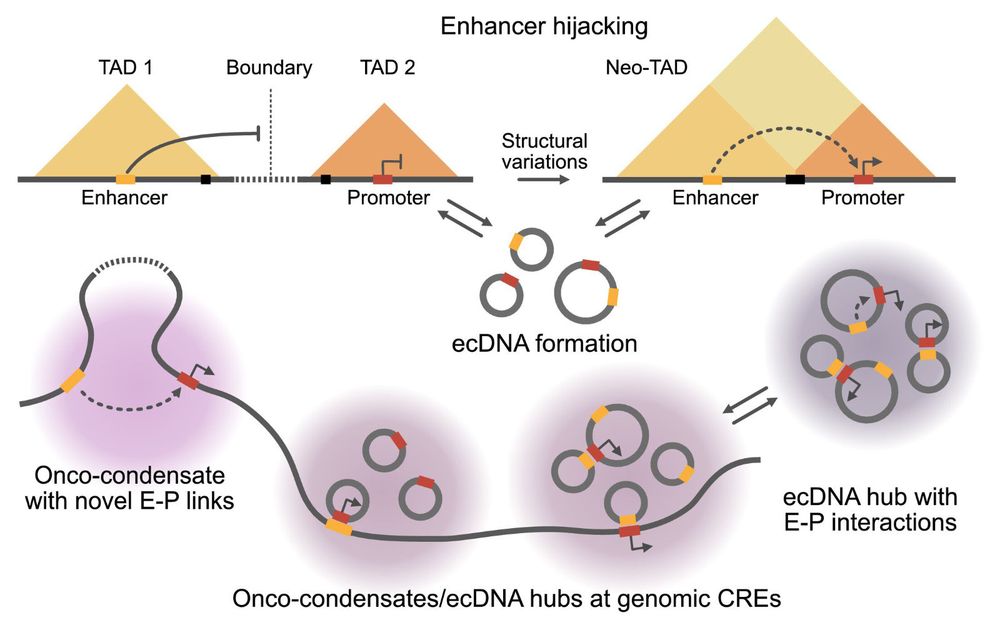
From enhancer hijacking to ecDNA and onco-condensates – we review how changes in nuclear architecture drive oncogenic gene expression by disrupting enhancer–promoter communication. doi.org/10.1002/ijc..... Kudos to Isabelle Seufert, @claire-vrgs.bsky.social and Sina Wille 👏
12.04.2025 20:38 — 👍 23 🔁 8 💬 1 📌 1

Trump Administration Has Begun a War on Science, Researchers Say (Gift Article)
Nearly 2,000 scientists urged that Congress restore funding to federal agencies decimated by recent cuts.
“We all rely on science.” U.S. scientists are speaking out for scientific integrity and academic freedom. What they’re doing matters. We owe them our solidarity and a clear stand beside them.
www.nytimes.com/2025/03/31/s...
31.03.2025 22:39 — 👍 13 🔁 6 💬 1 📌 1
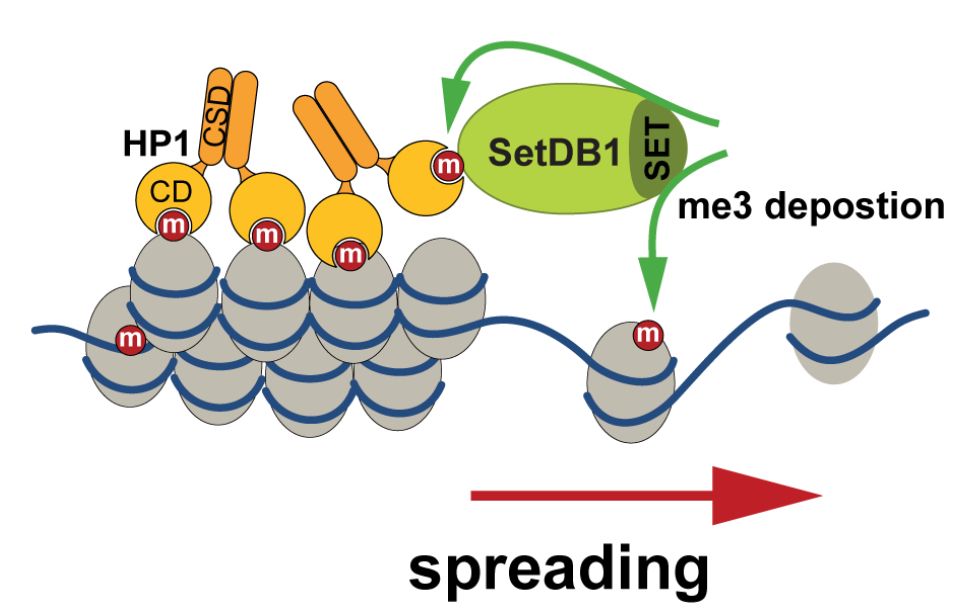
A mechanism for maintaining and spreading H3K9me3 in heterochromatin from the Fejes Toth and Aravin labs that depends on the local H3K9me3 density: HP1 dimers recruit SetDB1 to chromatin by simultaneously binding H3K9me3 on histone H3 and auto-methylated SetDB1. www.biorxiv.org/content/10.1...
25.01.2025 09:16 — 👍 38 🔁 8 💬 0 📌 0
🙏 9/ Kudos to lead author Robin Weinmann, @udupaarjun.bsky.social and @janfabio.bsky.social for shaking up heterochromatin and cracking this puzzle! Thanks to @dfgpublic.bsky.social for funding us in the SPP2191 program.
13.12.2024 13:47 — 👍 3 🔁 1 💬 0 📌 0
💡 8/ Time to think different: HP1α isn't a global chromatin compaction machine, but a local transcriptional gatekeeper that strong activators can bypass. A new view on how heterochromatin does its job.
13.12.2024 13:47 — 👍 3 🔁 1 💬 1 📌 0

💻 7/ Our mathematical modeling showed it could be beautifully simple: Each repeat unit independently switches between silent/active states. No fancy mechanisms needed - basic ligand binding math explains everything we see, with no evidence for HP1α phase separation in our mouse fibroblasts.
13.12.2024 13:47 — 👍 2 🔁 1 💬 1 📌 0

🔍 6/ Looking up close with super-resolution microscopy revealed: HP1α sits in small ~50 nm clusters that persist during activation. Just good old chromatin binding at work, with additional HP1α molecules recruited through protein-protein interactions.
13.12.2024 13:47 — 👍 2 🔁 1 💬 1 📌 0
💭 5/ Another surprise: Chromatin can unpack without histone acetylation! In cells lacking HP1α/H3K9me3, VP16 can decondense chromatin and activate genes without enriching H3K27ac - revealing an alternative paths to open up chromatin.
13.12.2024 13:47 — 👍 4 🔁 1 💬 1 📌 0
🚫 4/ HP1α acts as a local transcriptional brake in two ways: complete silencing or partial attenuation of transcription. Weak activators like VP16 are blocked, while strong ones like VPR can overcome this barrier - shown both at chromocenters and using a reporter system.
13.12.2024 13:47 — 👍 2 🔁 1 💬 1 📌 0
💥 3/ The big surprise: Even when the strongest activator VPR decondenses chromatin and drives transcription, HP1α and H3K9me3 stay put! This challenges the dogma that these repressive chromatin features must be removed for gene activation.
13.12.2024 13:47 — 👍 4 🔁 1 💬 1 📌 0
Epigenetic Inheritance, Neuroscience & anything biology-related
https://www.odedrechavilab.com/
q.e.d: https://www.qedscience.com
Organizer of “The Woodstock of Biology”
TED: https://shorturl.at/myFTY
Huberman Lab Podcast: https://youtu.be/CDUetQMKM6g
A journal dedicated to publishing the latest advances across all areas of cell biology. Part of @natureportfolio.nature.com
nature.com/ncb/index.html
Scientist at DKFZ and NCT Heidelberg
#epigenetics #stemcells #leukemia #colorectalcancer #multiomics #precisiononcology
#oncofetal #reprogramming
#plasticity
www.translational-cancer-epigenomics.de
physicist, bioinformatics / biostats researcher at University of Heidelberg
Oncologist-scientist at Weill Cornell and New York genome center. Into somatic evolution, trees and words
Professor at the NYU School of Medicine (https://yanailab.org/). Co-founder and Director of the Night Science Institute (https://night-science.org/). Co-host of the 'Night Science Podcast' https://podcasts.apple.com/us/podcast/night-science/id1563415749
Director, Centre for Evolution and Cancer @cec-icr.bsky.social at @icr.ac.uk
Genomics, epigenetics, evolution, mathematical modelling, data science, bowel cancer
typing...
(also active on X as @cees_dekker)
From embryos to equations and back.
We study how physical principles shape life, from collective DNA–protein interactions to mesoscale processes within cells and tissues.
Tweets by Grill Lab members.
https://grill-lab.org
The Banito Lab at the Hopp Children Cancer Center (KiTZ), German Cancer Research Center (DKFZ) in Heidelberg studies soft-tissue sarcomas - from underlying molecular mechanisms to development of new animal models.
e:Med, the BMBF's Germany-wide research program on systems medicine.
https://www.sys-med.de/de/
Research group leader @German Cancer Research Center, reconstructing human development and disease using cell fate engineering
Physician Scientist
Director, Heidelberg Myeloma Center
Views are my own
🧬 Join our world-class courses, conferences, and workshops at the forefront of molecular life science and its applications. 👩🏻🔬 www.embl.org/events/ 🔬
Physicist modelling the complexity of eukaryotic cell's nucleus. University Medical Centre Göttingen
Die DFG ist die größte Forschungsförderorganisation und die Selbstverwaltungsorganisation der Wissenschaft in Deutschland.
www.dfg.de
Impressum: https://www.dfg.de/de/service/kontakt/impressum
Social Media-Netiquette: https://www.dfg.de/de/service/presse
Experimental and computational research group in regulatory, genome, and cancer evolution.
Currently (v2) at the DKFZ / Heidelberg University (2019-now)
Founded (v1) at the CRUK Cambridge Institute / University of Cambridge (2006-2022)
Life scientist.
Head of EMBO Membership & Elections and Courses & Workshops @embo.org #EMBOevents
previously: Editor at Molecular Systems Biology, EMBO Press
Views my own