
selfie of a sleepy biologist with his research poster on an insect lipid flippase
If you're around for the Just-B poster session today at #bps2026 Exhibition Halls, do drop by B-56 if you're interested in lipid transporters!
23.02.2026 21:23 —
👍 3
🔁 0
💬 0
📌 0
Incredibly excited to share our findings on the fruit fly lipid flippase! Please join the #bps2026 poster session at 1.45 PM today and the Membrane Transporters Platform on Tuesday! Big thanks to the organisers for their recognition and @sandipbasak.bsky.social for his generous guidance.
22.02.2026 19:46 —
👍 2
🔁 0
💬 0
📌 0
An impressive number of graduate applications this year used the phrase “curiosity-driven approach” 🤔
17.01.2026 22:06 —
👍 597
🔁 51
💬 9
📌 19

Asymmetric gating of a homopentameric ion channel GLIC revealed by cryo-EM | PNAS
Pentameric ligand-gated ion channels (pLGICs) are vital neurotransmitter receptors
that are key therapeutic targets for neurological disorders. Alt...
We are excited to share our latest work on GLIC published in @pnas.org, where we developed a #cryoEM data processing strategy that allowed us to isolate and reconstruct multiple asymmetric states, revealing detailed insights into the channel’s activation mechanism. tinyurl.com/mwtb4rpj
24.10.2025 05:31 —
👍 16
🔁 6
💬 1
📌 0
It is like magic. No one really wants to know how a trick is done. they want to believe it is real. With LLM they are more interested in talking about what it MIGHT do, or what they think it can do. Then what it actually does.
18.12.2025 04:21 —
👍 26
🔁 9
💬 0
📌 0
thanks for sharing this
17.11.2025 06:31 —
👍 1
🔁 0
💬 0
📌 0

Atkinson Hyperlegible Font - Braille Institute
Read easier with Atkinson Hyperlegible Font, crafted for low-vision readers. Download for free and enjoy clear letters and numbers on your computer!
periodic reminder of the existence of Atkinson Hyperlegible, a free font available from the Braille Institute designed to improve readability for people with low vision
I use it in talks because it's pretty and also because, as an audience member, I am perpetually squinting at people's slides
17.11.2025 04:19 —
👍 650
🔁 323
💬 19
📌 19

the horrors persist, but so DOI (a doi is a persistent identifier for academic papers, e.g. 10.1000/182)
#academicsky #librarysky #libsky #skybrarians #medlibs do u guys like persistent identifier memes?
22.03.2025 23:07 —
👍 139
🔁 33
💬 8
📌 4
OpenAI doesn't report a single number for its climate impacts - not anywhere, in any way, shape or form.
It sounds kind of obvious but the fact that a company with such incredible electricity hunger isn't disclosing basic information about its energy consumption and associated emissions is v bad..
28.12.2024 07:58 —
👍 5839
🔁 1540
💬 91
📌 61
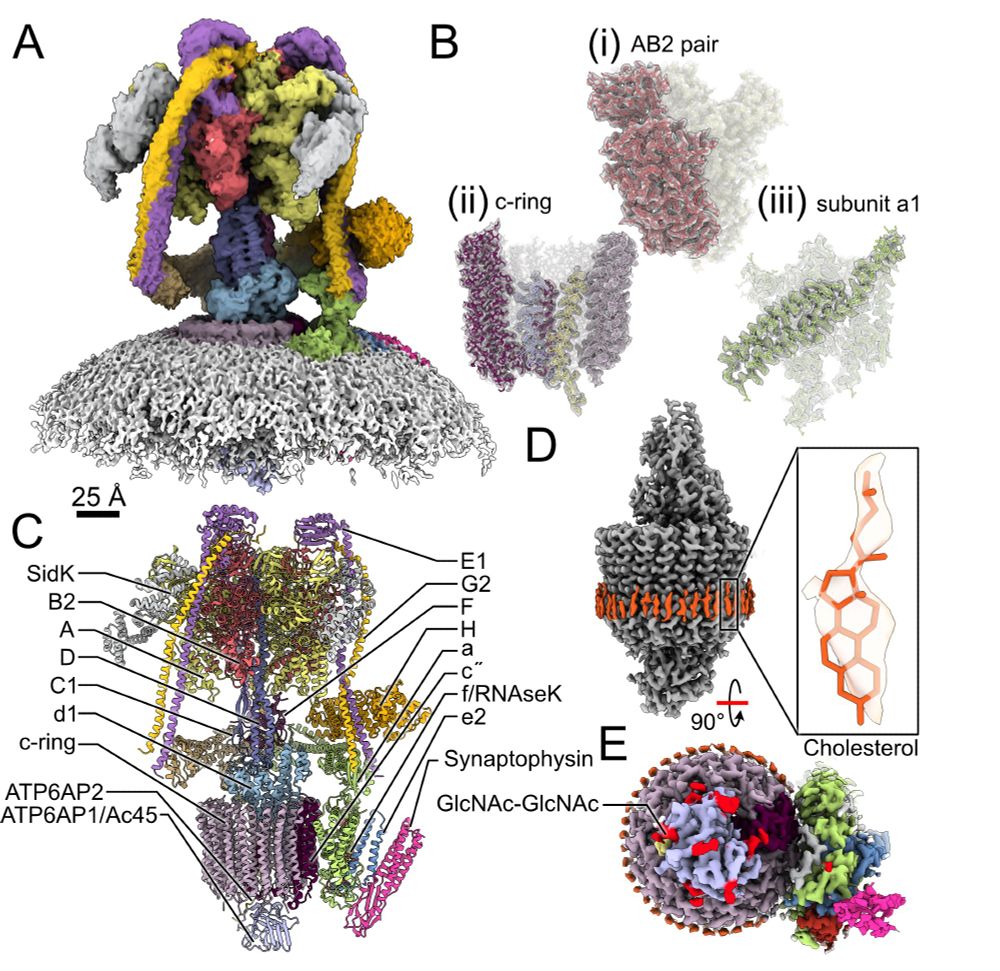
Years after the idea and preliminary experiments, Claire Coupland's amazing structure of the endogenous vacuolar- or vesicular-ATPase in its NATIVE vesicle is now in bioRxiv "High resolution cryo-EM of V-ATPase in native synaptic vesicles"
www.biorxiv.org/content/10.1...
02.04.2024 17:11 —
👍 119
🔁 39
💬 6
📌 4
im on every app. im in your city, in your neighborhood. the sales associate who handed you a large when you asked for a medium? that was me. i am now behind you.
20.11.2024 07:00 —
👍 34688
🔁 2739
💬 575
📌 269
yes, thanks for the discussion!
20.11.2024 08:00 —
👍 1
🔁 0
💬 0
📌 0
this is also correctly predicted
20.11.2024 07:55 —
👍 0
🔁 0
💬 1
📌 0
yep correct, the small protein is the green box in bottom right. This is a large membrane protein. The interacting region of the first protein can be verified by homology modelling. All the high confidence regions predicted by AF3 can be verified by homology, and inter domain packing is also good.
20.11.2024 07:54 —
👍 0
🔁 0
💬 0
📌 0
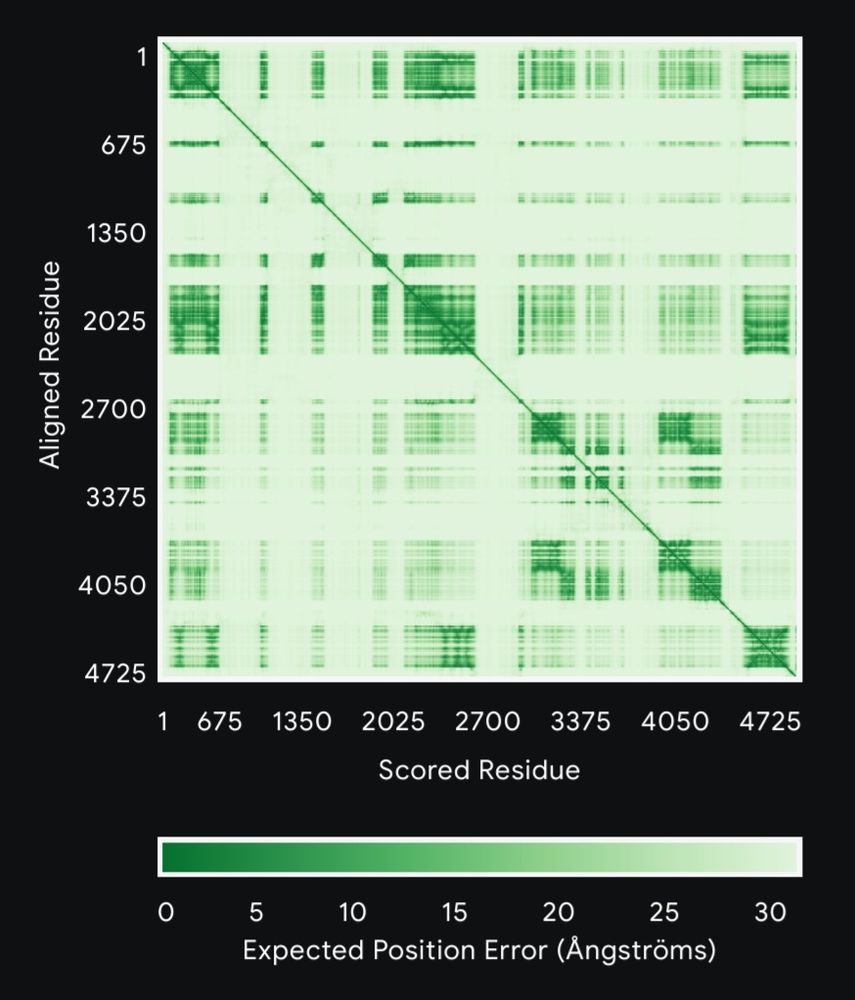
PAE plot from AF3
PAE looks like this. The iPTM score is also 0.74, so these helices are really unreliable, and they take up a considerable amount of amino acids in the total sequence.
20.11.2024 07:32 —
👍 1
🔁 0
💬 1
📌 0
ah I see, thanks
20.11.2024 07:12 —
👍 0
🔁 0
💬 0
📌 0
thanks, yeah I figured because manual inspection of the sequence and IDR prediction server also spit out similar disordered behaviour for these regions. Most of these are just poly-Ns, some need to be checked more carefully.
20.11.2024 06:45 —
👍 0
🔁 0
💬 2
📌 0
ohh that low confidence in the middle of the curl..yikes.
20.11.2024 06:33 —
👍 0
🔁 0
💬 0
📌 0
ohh do you have a visual example of 'curled' models?
20.11.2024 05:21 —
👍 0
🔁 0
💬 1
📌 0
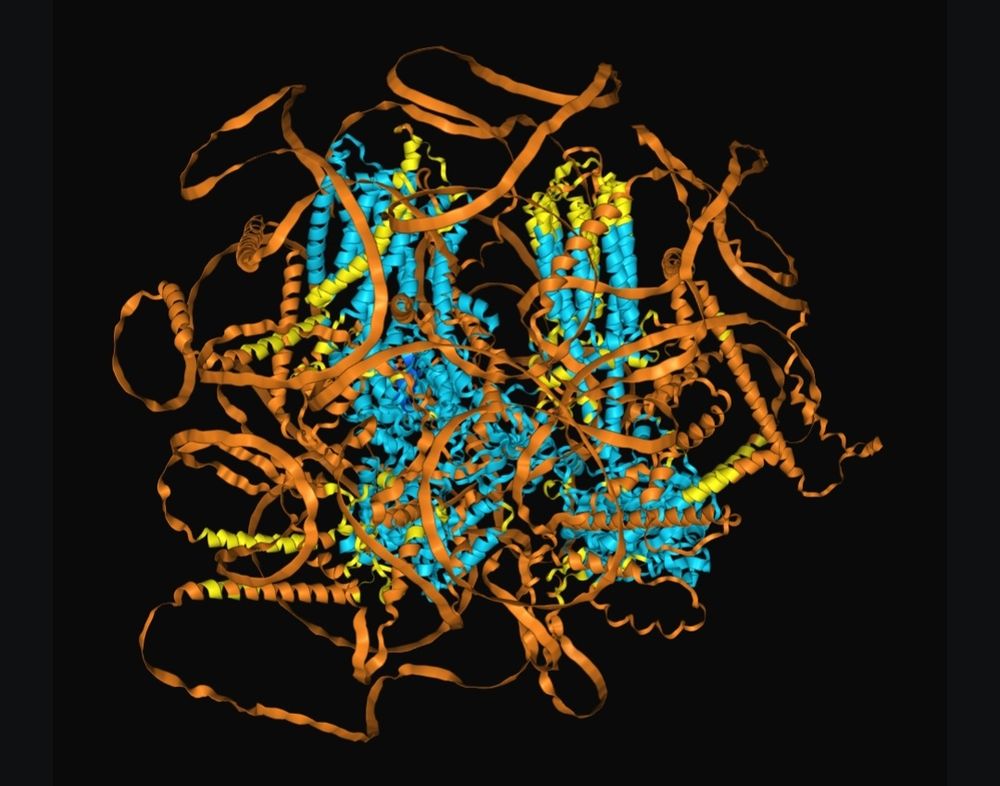
A 500 kDa protein model generated by AlphaFold 3
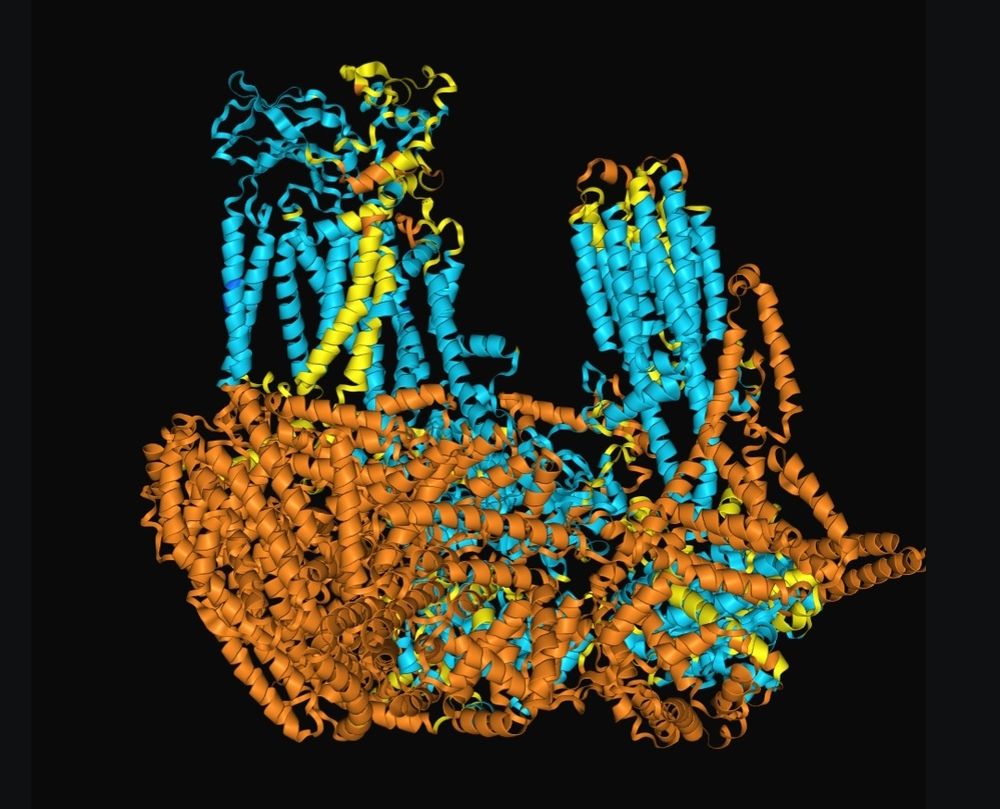
AF3 model of the protein with the binding partner
AF3 predictions for a 500 kDa protein without (left) and with (right) a known 40 kDa binding partner. Apparently disordered regions collapsing into low confidence cytosolic helices. Is AF hallucinating these helices?
19.11.2024 13:43 —
👍 16
🔁 2
💬 6
📌 1
thanks!
17.11.2024 12:07 —
👍 1
🔁 0
💬 0
📌 0
🙌🙌
17.11.2024 12:04 —
👍 1
🔁 0
💬 1
📌 0
haha it is art as long as it provokes a human emotion, pain in this case
15.11.2024 12:20 —
👍 2
🔁 0
💬 0
📌 0
yep, it was apparently a fabrication issue because we didn't notice anything strange while handling and preparing this grid, it was only clear at the screening stage
15.11.2024 12:19 —
👍 1
🔁 0
💬 1
📌 0
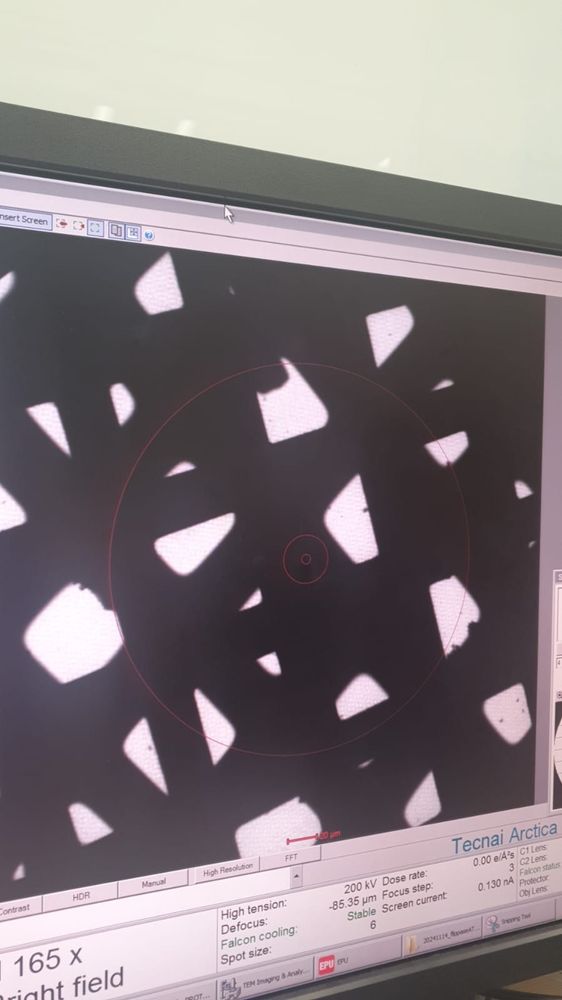
bad cryoEM grid imaged at low mag
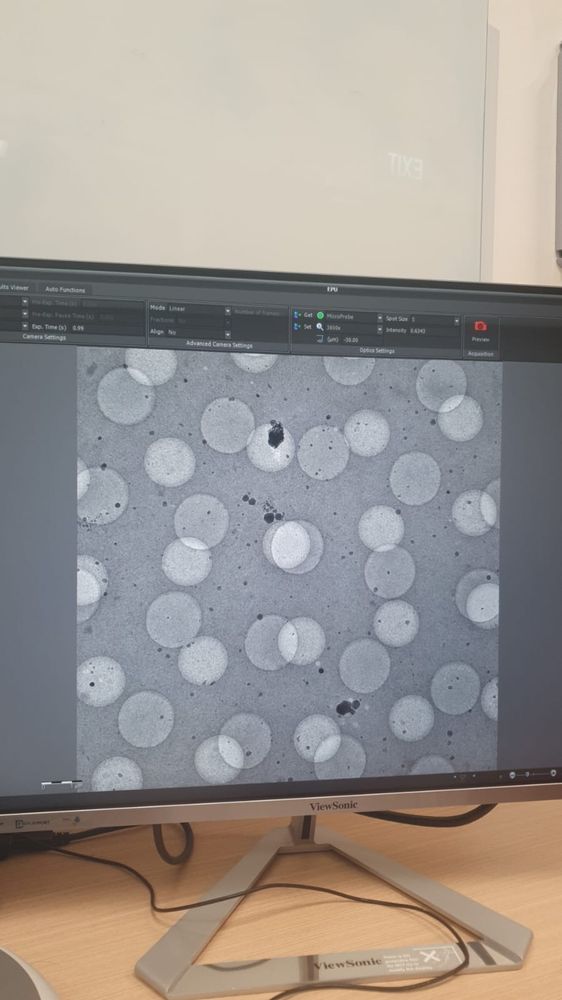
bad cryoEM grid imaged at high mag
pain, just literal pain
15.11.2024 11:34 —
👍 7
🔁 0
💬 4
📌 0
trying to fix the student numbers right here 😬 hopefully my friends pop in too
13.11.2024 14:31 —
👍 2
🔁 0
💬 0
📌 0
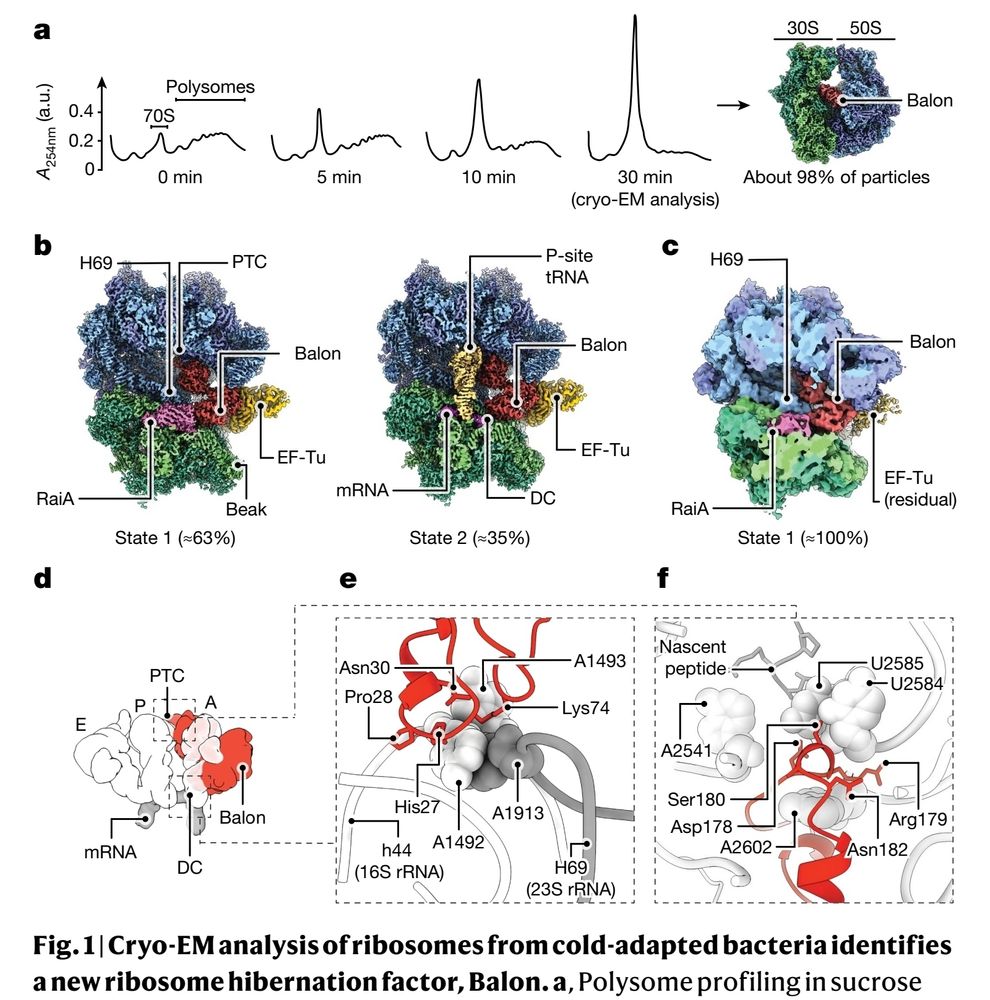
Been thinking about creating a collection of good protein structure figures, as inspiration for my own work.
#1 www.nature.com/articles/s41...
03.03.2024 08:37 —
👍 173
🔁 54
💬 7
📌 10











