On a personal note, it is at once surreal, gratifying and humbling to be part of a wet-dry colab, I’m so grateful for my collaborators (also great friends) for making it real + keeping it fun! Also thankful our buildings (BAIR and @innovativegenomics.bsky.social) are right next to each other :)
31.05.2025 15:48 — 👍 2 🔁 0 💬 0 📌 0
This work is only made possible through an incredible interdisciplinary collaboration between the Listgarten lab and @savagecatsonly.bsky.social . All kudos go to the amazing team that I’m super grateful to be part of: @hnisonoff.bsky.social @marialukarska.bsky.social (and Ishan and Luke)
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0
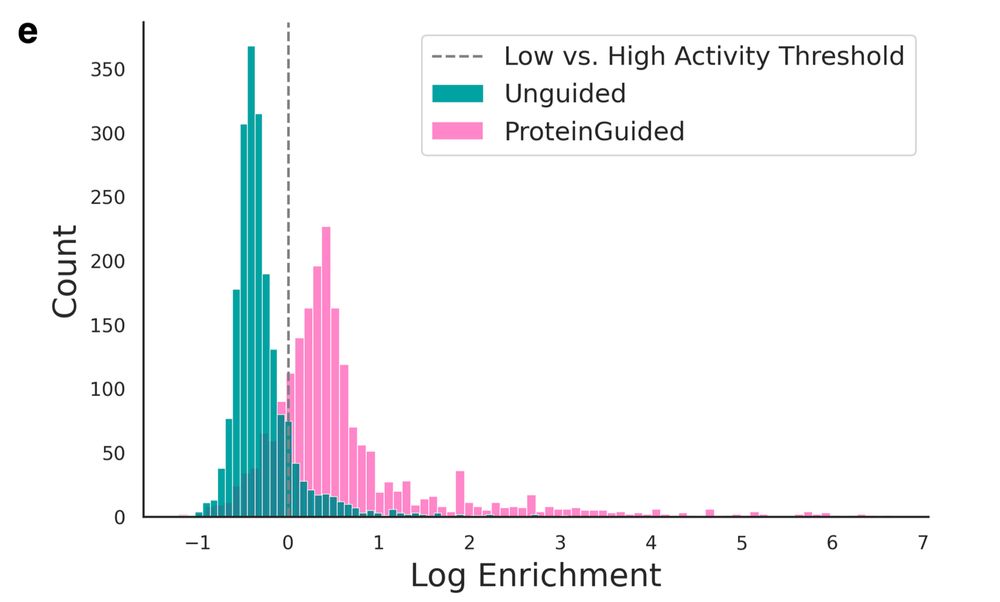
The guided library in round 2 showed significantly higher activity than the initial unguided library in the experimental base editing assay.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0

We didn't just validate in silico - we also synthesized & tested proteins in the lab. We used ProteinGuide to engineer an adenine base editor for high activity: generated 2,000 variants → tested in bacteria → used results to guide 2,000 new designs.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0

In our third task, we demonstrate the generality of ProteinGuide beyond amino acid sequences, to structure tokens. In particular, we guide ESM3 to generate backbone structures (as tokens) with specified CATH fold class labels.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0

In our second task, we guided ESM3 to re-design enzymes sequences predicted to belong to specific enzyme classes, based on a published classifier, CLEAN, for enzyme commission number.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0

In our first task, we guided ProteinMPNN with experimental stability measurements from the @grocklin.bsky.social lab to generate amino acid sequences encoding proteins that are more stable than what ProteinMPNN would do on its own.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0
To illustrate the potential of ProteinGuide, we applied it, in silico, to three tasks, using two representative, well-known protein generative models, ProteinMPNN and ESM3. Across these three tasks, we observed that guidance, as expected, led to the desired outcome.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0

We leverage the fact that MLMs (e.g., ESM3), OA-AR models (e.g., ProteinMPNN), and masking-based diffusion models are actually equivalent. This allows us to leverage our previously-developed guidance methodology for discrete diffusion and flow models for MLMs and OA-AR models.
31.05.2025 15:45 — 👍 1 🔁 0 💬 1 📌 0
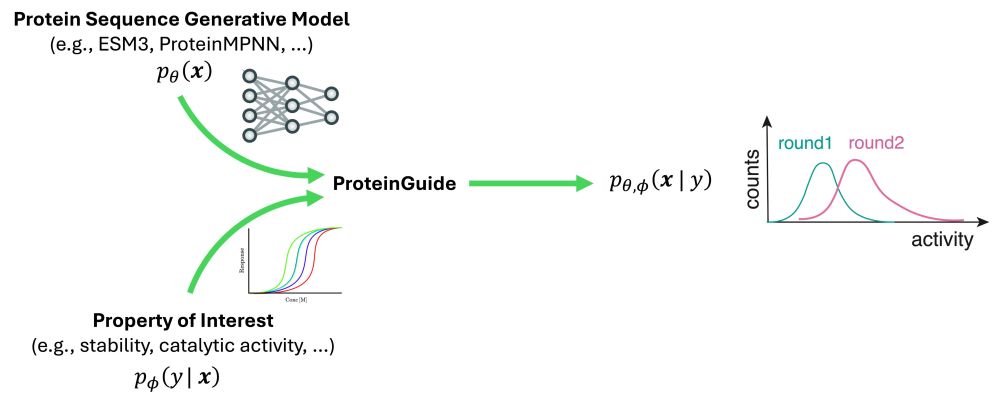
Guide your favorite protein generative model with experimental data? Meet ProteinGuide - a method to condition pre-trained models on properties without retraining. We validated it both in silico by guiding ProteinMPNN and ESM3 on 3 tasks and in vitro by engineering base editors.
31.05.2025 15:45 — 👍 11 🔁 5 💬 1 📌 2
Reporting from the frontiers of health & medicine.
Assistant Professor, University of Washington, Genome Sciences.
Previous: JSMF Fellow, Berkeley EECS
♡: Computational biology, evolutionary dynamics, quantitative immunology
https://dewitt-lab.github.io/
[disclaimer: opinions mine]
Assistant Professor in EEMB @ UCSB | Evo-Devo • Botany • phylogenetic comparative methods • meristems
https://www.martinezgomezlab.com/
Structural Biologist turned computational biologist @UCBerkeley. Former Miller Fellow
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Using genome engineering to solve humanity’s greatest problems in health, climate & sustainable agriculture. UC Berkeley, UCSF, UC Davis. https://innovativegenomics.org/
Compartmentalizing your metabolism, and y’know, CRISPR stuff. Account managed by grad students and postdocs 🤙
@ucberkeleyofficial.bsky.social @innovativegenomics.bsky.social @hhmi.bsky.social
savagelab.org
Research Director, Founding Faculty, Canada CIFAR AI Chair @VectorInst.
Full Prof @UofT - Statistics and Computer Sci. (x-appt) danroy.org
I study assumption-free prediction and decision making under uncertainty, with inference emerging from optimality.
Machine learning prof at U Toronto. Working on evals and AGI governance.
Machine Learning Professor
https://cims.nyu.edu/~andrewgw
Scientist, Assistant Professor at MIT biology, #FirstGen
PhD student at UC Berkeley
Machine Learning for Protein Engineering
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app







