Such an absolute honour 🙏 Thank You @bscb-official.bsky.social Incredibly lucky to be surrounded by the most supportive colleagues and community. This award truly belongs to my wonderful, wonderful lab @crick.ac.uk
22.01.2026 11:14 — 👍 63 🔁 5 💬 16 📌 1
🚨Do sign up to our mailing list for this very exciting joint talk by Profs Marino Arroyo and Xavier Trepat on active morphogenesis on Wednesday 21st Jan🚨
19.01.2026 09:43 — 👍 4 🔁 3 💬 0 📌 0
8. We thank team member Zuzhi Jiang, collaborators Binyamin Zuckerman, Leor Weinberger, Matt Thomson, funding sources @cancerresearchinst (@CancerResearch) @NIH @czbiohub @czi @UCSF, the thoughtful reviewers from @NatureBiotech, and everyone else who shared data and feedback!
13.01.2026 13:08 — 👍 0 🔁 0 💬 0 📌 0
GitHub - Gartner-Lab/Concord
Contribute to Gartner-Lab/Concord development by creating an account on GitHub.
7. Best of all, it's fast (1M cells in <10 mins) and memory-friendly.
💻 Open Source: github.com/Gartner-Lab/... 📖 Docs: qinzhu.github.io/Concord_docu... Try it out and let us know what you think!
13.01.2026 13:08 — 👍 1 🔁 0 💬 1 📌 0

modalities
6. It's not just for scRNA-seq. CONCORD seamlessly handles and aligns scATAC-seq and spatial transcriptomics (Xenium, etc.), revealing fine-grained subtypes across technologies
13.01.2026 13:08 — 👍 0 🔁 0 💬 1 📌 0

intestinal differentiation
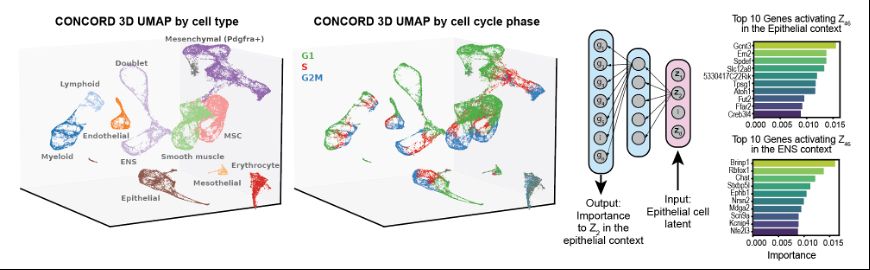
5. CONCORD resolved complex topologies during intestinal development, such as differentiation trajectories linked to cell cycle loops and providing biologically interpretable encodings traced to gene co-expression programs (click here for interactive 3D UMAP! qinzhu.github.io/Concord_docu...):
13.01.2026 13:08 — 👍 2 🔁 0 💬 1 📌 0

lineage detail
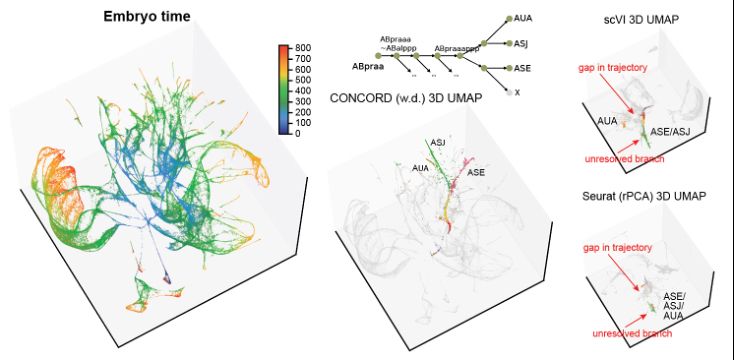
4. On a whole-organism, multi-species embryonic atlas of C. elegans and briggsae from @jisaacmurray’s lab where lineage annotations provide a ground truth, CONCORD resolved lineage trajectories and bifurcations (see animation and click here for interactive 3D UMAP! qinzhu.github.io/Concord_docu...):
13.01.2026 13:08 — 👍 1 🔁 0 💬 1 📌 0

benchmarking
3. With this sampling scheme, we found that even a minimalist single-layer network achieves better-than-SOTA performance. It disentangles biology from batch effects without over-correcting.
We benchmarked it rigorously. Across a diverse set of datasets, CONCORD consistently ranked top:
ben
13.01.2026 13:08 — 👍 3 🔁 0 💬 1 📌 0

minibatch sampler overview
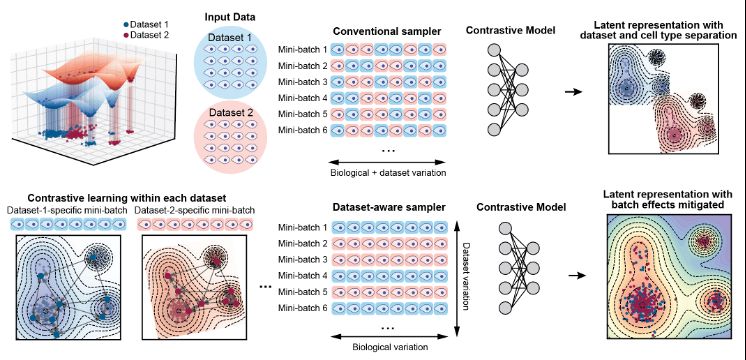
2. At the core is a novel mini-batch sampler that changes how the model 'sees' data. Unlike uniform sampling, our approach is two-fold: 🔹 Dataset-aware: Extracts consistent biological structure across batches. 🔹 Hard-negative: Distinguishes differences between closely related cell states.
13.01.2026 13:08 — 👍 0 🔁 0 💬 1 📌 0
1. Excited to share CONCORD, out in @NatureBiotech, an ML framework for single-cell analysis addressing integration, dimensionality reduction, and denoising in one go by @qinzhu1.🔗 www.nature.com/articles/s41... Check out this CONCORD model of worm development resolving differentiation trajectories:
13.01.2026 13:08 — 👍 13 🔁 4 💬 1 📌 0

Mind blown 🤯
25.10.2025 16:02 — 👍 0 🔁 0 💬 0 📌 0

We are all super happy and proud to see our work on the function and evolution of the #cephalic #furrow published in @nature.com. Let me say a few things about the background and history of this work on the #Evolution_of_Morphogenesis (1/12)
04.09.2025 08:21 — 👍 347 🔁 118 💬 16 📌 8
Very excited to be co-organizing this meeting next February. Fellow collective behavior enthusiasts, this one is for you!
21.08.2025 12:04 — 👍 11 🔁 6 💬 0 📌 0
Aria Huang's @azyhuang.bsky.social paper is out on beautiful embryonic kidney 3D cultures that branch properly and enable live defect analysis. She also finds intriguing effects of adhesion and stiffness on explant size, shape, and nephron formation - boundary conditions matter!
rdcu.be/eBSUK
22.08.2025 15:46 — 👍 32 🔁 11 💬 0 📌 2
🔔 Proud to share the preprint of my PhD work in the Petridou group @nicolettapetridou.bsky.social @embl.org ✨
“A closed feedback between tissue phase transitions and morphogen gradients drives patterning dynamics” 🐟 🔁 📶
🔗 www.biorxiv.org/content/10.1...
#devbio #biophysics
🧵⤵️
11.06.2025 16:47 — 👍 100 🔁 31 💬 3 📌 9

Workshop
New Technologies for Studying and Reprogramming Development
Organisers: Alex Dunn, Zev Gartner, Adrian Jacobo and Matthew Kutys
Early-career researchers apply for funded places
16-19 November 2025
Buxted Park, East Sussex, UK
One month left for ECRs to apply for a funded place at our Workshop 'New Technologies for Studying and Reprogramming Development' organised by Alex Dunn,
@zevgartner.bsky.social , @adrianjacobo.bsky.social and Matthew Kutys.
📆Apply by 23 May
biologists.com/workshops/no...
#BiologistsWorkshops
23.04.2025 13:34 — 👍 5 🔁 5 💬 0 📌 0

Background of DNA strand with logos of King's College London and the Leverhulme Trust and text Force Talks Mechanics of Life Leverhulme DSP seminar series Dr Jean-Leon Maitre Institut Curie Mechanics of blastocyst morphogenesis Wednesday, 07 May 2025, 15:00-16:00 email:mechanicsoflife@kcl.ac.uk for more information
Join us for our next #ForceTalk on "Mechanics of blastocyst morphogenesis" with Dr Jean-Leon Maitre @maitrejl.bsky.social of Institut Curie:
🗓️ 7 May 2025
🕒 15:00 - 16:00 BST
Online and open to all, link to join 👉 www.kcl.ac.uk/events/force...
#mechanobiology @kingsnmes.bsky.social
25.04.2025 11:15 — 👍 8 🔁 7 💬 1 📌 1

Workshop
New Technologies for Studying and Reprogramming Development
Organisers: Alex Dunn, Zev Gartner, Adrian Jacobo and Matthew Kutys
Early-career researchers apply for funded places
16-19 November 2025
Buxted Park, East Sussex, UK
Early-career researchers can apply for a funded place at our Workshop 'New Technologies for Studying and Reprogramming Development' organised by Alex Dunn,
@zevgartner.bsky.social , @adrianjacobo.bsky.social and Matthew Kutys.
📆Apply by 23 May
biologists.com/workshops/no...
#BiologistsWorkshops
11.04.2025 10:43 — 👍 18 🔁 11 💬 0 📌 1
Since I started my PhD in 2013, I have been tantalised by a question: can we measure or evaluate the transcriptional and mechanical states of cells in a tissue at the same time? I pursued this question as a postdoc and then as a PI! A🧵on the answer we have found 👇:
17.03.2025 12:52 — 👍 4 🔁 1 💬 1 📌 0
GitHub - Gartner-Lab/Concord
Contribute to Gartner-Lab/Concord development by creating an account on GitHub.
9. While we focused on scRNA-seq, early results suggest CONCORD’s applicability to spatial transcriptomics & scATAC-seq. It’s open-source in Python. See galleries: qinzhu.github.io/Concord_docu.... Try it: github.com/Gartner-Lab/....
20.03.2025 18:08 — 👍 2 🔁 0 💬 0 📌 0
8. CONCORD integrates datasets based solely on gene co-expression structures—without assuming batch-effect models or shared cellular states. It achieves robust alignment even with minimal dataset overlap, offering faster and scalable analysis from small studies to atlas-scale projects.
20.03.2025 18:08 — 👍 1 🔁 0 💬 1 📌 0
neighborhood and dataset aware sampling improves learning
7. The key innovation leading to this performance gain is a probabilistic, dataset- and neighborhood-aware sampling strategy tailored for self-supervised contrastive learning, enabling the generation of coherent, denoised and high-resolution latent cell representations across diverse datasets:
20.03.2025 18:08 — 👍 2 🔁 0 💬 1 📌 0
6. CONCORD also provides biologically interpretable encodings representative of context-dependent gene-expression programs.
20.03.2025 18:08 — 👍 0 🔁 0 💬 1 📌 0
loops, cylinders and filaments in a gut developmental atlas
5. An intestinal development atlas demonstrated CONCORD's ability to resolve complex topologies, including differentiation trajectories intertwined with cell cycle loops (click here for interactive 3D UMAP! qinzhu.github.io/Concord_docu...):
20.03.2025 18:08 — 👍 1 🔁 0 💬 1 📌 0
see lineage relationships in C. elegans developmental data sets
4. We validated CONCORD on an embryonic atlas of C. elegans and C. briggsae from @jisaacmurray. CONCORD captured known bifurcations, resolved subtle cell states, and uncovered lineage convergence events missed by other methods (click here for interactive 3D UMAP! qinzhu.github.io/Concord_docu...):
20.03.2025 18:08 — 👍 2 🔁 0 💬 1 📌 0
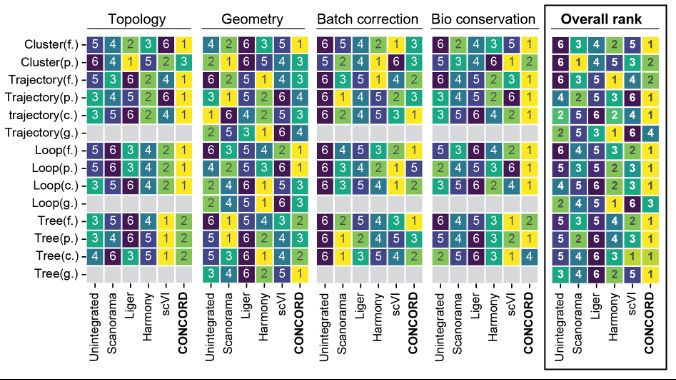
CONCORD outperforms other methods on metrics of geometry and topology
3. We evaluated CONCORD on datasets with topologies such as continuous trajectories, loops, clusters and hierarchical trees. Using scIB metrics, geometric measures, and TDA, we assessed its ability to preserve biological structure. CONCORD outperforms existing methods:
20.03.2025 18:08 — 👍 0 🔁 0 💬 1 📌 0
PhD student in the Fre lab at @institutcurie.bsky.social
Tissue morphogenesis & Fate specification in epithelia.
Organoid and live imaging.
Alumni AgroParisTech & ENS Ulm
A Mathematician dabbling in Data Science, especially unsupervised learning and data exploration. UMAP, HDBSCAN, PyNNDescent, DataMapPlot. (He/Him)
Postdoc, Hughes Lab @ UPenn | PhD, Shvartsman Lab @ Princeton
Part-time physicist-turned-biologist studying early embryos and the cell cycle @KU Leuven, full-time dad to two active boys. Visualizing academic data trends in Flanders.
www.gelenslab.org | https://academic-compass.be/en/
A genomics cat, sometimes Christopher H. Browne Professor of Biology at Penn. All posts my opinion only.
zookeeper to two tiny humans, professor, cell herder, bioelectrician, 'professional' storyteller, and waterbears just because. see us at:
cohenlab.princeton.edu
Theoretical Biological and Soft Matter Physicist. Interested in all things networks-related in biological physics and physical biology. My views here are my own.
Husband, Father, Grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science, Stand Up for Science advisor, Shenanigator, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Scientist and mother…passionate about traveling and embracing challenges.
Dedicated to unraveling the mysteries of multicellular systems and stem cell biology. Combining experimental and theoretical approaches to understand the dynamics of cell behaviour
Computational Biology @ETH: data-driven modeling & simulation of emerging phenomena in development & disease https://bsse.ethz.ch/cobi https://youtube.com/@cobi-ethz
#interdisciplinary Science project - A*MIDEX (AMU)
- #Research #Education #Engineering
Lab-run account @ Penn Bioengineering. Studying development and tissues using micro-scale engineering, chemical biology, and microscopy. alexhugheslab.com
Assistant Professor at Caltech | Chemical Biologist
morsteinlab.caltech.edu
The choreography of multicellular systems, computer simulations, gene regulatory circuits, and swarm robotics.
Head of EMBL Barcelona
Co-chair of the Barcelona Collaboratorium
International research centre devoted to curing #cancer and other diseases linked to #aging. @cerca.cat center, member of @bist.eu.
www.irbbarcelona.org
Scientist studying development of embryos and building their models from stem cells. The author of #TheDanceofLife. Warsaw/Oxford/Cambridge/LA.
Science artist and cancer researcher. Huntsman Cancer Institute, University of Utah
www.bryanwelm.com
Chemist at HHMI's Janelia Research Campus. Passionate about designing, building, and giving away fluorescent dyes to illuminate biological systems. Striving to be positive about all things chemistry (except ChemDraw).
ORCID: 0000-0002-0789-6343





















