
Uptake of anti-cancer drugs depends on N-terminally acetylated anion channels | @ispt-proteinterm.bsky.social www.nature.com/articles/s42...
07.10.2025 11:03 — 👍 1 🔁 1 💬 0 📌 0@ispt-proteinterm.bsky.social
Non-profit society of scientists devoted to understanding the molecular mechanisms and biological role of protein termini in plants, animals, fungi and bacteria. Conference every second year. #acetylation #arginylation #oxidation #N-degron #C-degron

Uptake of anti-cancer drugs depends on N-terminally acetylated anion channels | @ispt-proteinterm.bsky.social www.nature.com/articles/s42...
07.10.2025 11:03 — 👍 1 🔁 1 💬 0 📌 0
A Cell-Potent Bisubstrate Inhibitor to Probe NatD Acetyltransferase Activity | The inhibitor effectively reduced cellular Nα-acetylation on histone H4, leading to reduced migration in lung cancer cells | @acs.org ACS Chemical Biology @ispt-proteinterm.bsky.social pubs.acs.org/doi/10.1021/...
05.10.2025 12:06 — 👍 4 🔁 2 💬 0 📌 0
Protein N-terminal acetylation regulates the growth of rice plants | #NAA30 #NatC #rice #acetylation www.sciencedirect.com/science/arti...
25.09.2025 09:45 — 👍 2 🔁 2 💬 0 📌 0Two new volumes of Methods in Enzymology with 29 chapters/papers on Protein termini methods and reviews are now out! Thx to all authors!🙌 @ispt-proteinterm.bsky.social #N-degron #prenylation #myristoylation #formylation #arginylation #oxidation #acetylation www.sciencedirect.com/bookseries/m...
25.09.2025 09:17 — 👍 4 🔁 2 💬 1 📌 0
Developing leaves form a spatiotemporal oxygen gradient that is sensed through the oxygen sensing machinery and directs morphogenesis | New preprint from @daanweits.bsky.social and @syno2xis.bsky.social labs #N-degron #Cysteine #hypoxia @ispt-proteinterm.bsky.social www.biorxiv.org/content/10.1...
23.09.2025 07:38 — 👍 4 🔁 2 💬 1 📌 0
Anatomy and habitat shape the oxygen sensing machinery of flowering plants | new preprint from Licausi lab @syno2xis.bsky.social #hypoxia #N-degron @ispt-proteinterm.bsky.social www.biorxiv.org/content/10.1...
18.09.2025 12:40 — 👍 8 🔁 5 💬 1 📌 0
New review on the generation and consequences of N-terminal proteoform diversity from Evan Morrison and Olivia Rissland | #N-degron #RNA @cp-cellreports.bsky.social www.sciencedirect.com/science/arti...
17.09.2025 08:09 — 👍 3 🔁 3 💬 0 📌 0
Targeted degradation of pathogenic TDP-43 proteins in amyotrophic lateral sclerosis using the AUTOTAC platform | Yong Tae Kwon lab | #N-degron www.biorxiv.org/content/10.1...
08.09.2025 11:07 — 👍 8 🔁 4 💬 0 📌 0
A synthetic ERFVII-dependent circuit in yeast sheds light on the regulation of early hypoxic responses of plants | Preprint from @mikellavi.bsky.social @beagiuntoli.bsky.social @syno2xis.bsky.social #N-degron #hypoxia www.biorxiv.org/content/10.1...
08.09.2025 10:15 — 👍 7 🔁 4 💬 0 📌 0Want to know how lipidation of some nascent chains takes place by NMT2-- Check our latest work on how NAC couples Protein Synthesis with Nascent Polypeptide Myristoylation on the Ribosome out today @embojournal.org: www.embopress.org/doi/full/10....
26.08.2025 11:00 — 👍 42 🔁 21 💬 2 📌 2
Nitric oxide promotes cysteine N-degron proteolysis through control of oxygen availability | Important finding by Kim, Tian, Ratcliffe and Keeley @ox.ac.uk @ispt-proteinterm.bsky.social #N-degron www.pnas.org/doi/10.1073/...
20.08.2025 10:39 — 👍 10 🔁 8 💬 0 📌 0
Disease resistance in rice steered by a long non-coding RNA via protein N-terminal acetylation | @ispt-proteinterm.bsky.social advanced.onlinelibrary.wiley.com/doi/10.1002/...
19.08.2025 12:51 — 👍 3 🔁 1 💬 0 📌 0
Melon wrinkles linked to protein N-terminal acetylation. link.springer.com/article/10.1...
19.08.2025 12:19 — 👍 1 🔁 1 💬 0 📌 0
Global profiling of N-terminal cysteine–dependent degradation mechanisms | PNAS | @ispt-proteinterm.bsky.social www.pnas.org/doi/10.1073/...
18.08.2025 13:08 — 👍 8 🔁 2 💬 0 📌 0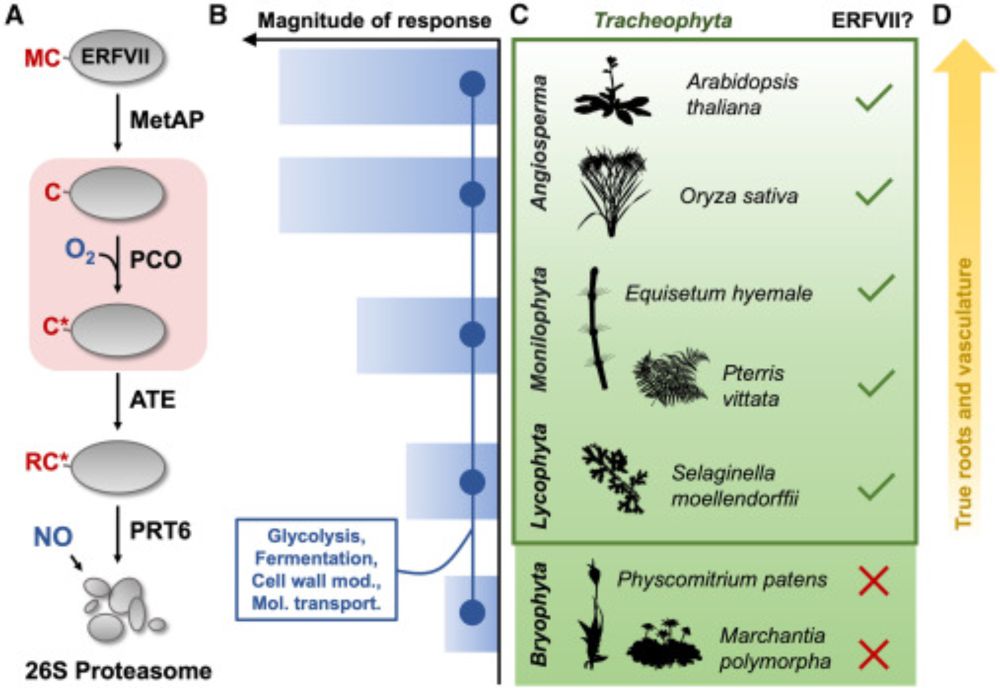
Our 'Spotlight' discussing an excellent recent paper by @lauradc.bsky.social @n-end-rules.bsky.social & @syno2xis.bsky.social et al on the evolution of oxygen signalling in land plants is now online at Molecular Plant.
Spotlight: tinyurl.com/7ydn6p4a
Original paper: tinyurl.com/yw5yd4f8

📢 Our August issue is online!
🎨 The cover highlights a Feature Review from Stephanie Rosswag de Souza, @gzaffagnini.bsky.social, and @bokelab.bsky.social on how proteostasis functions in cellular dormancy across a number of examples, from yeast to adult quiescent stem cells and vertebrate oocytes.
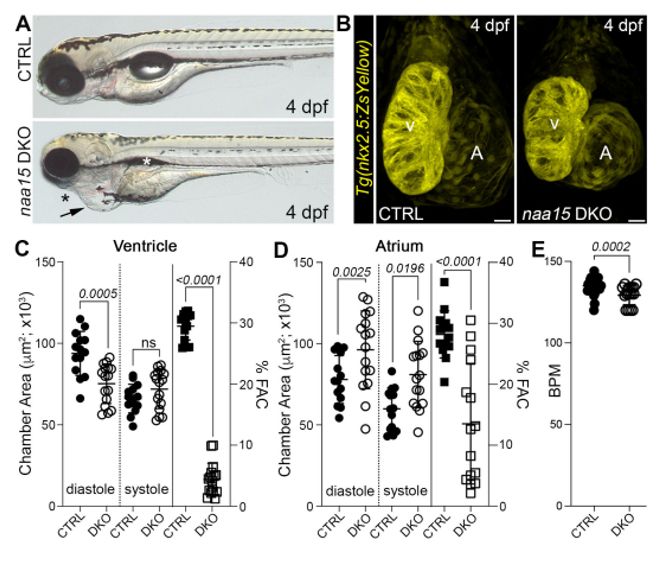
Zebrafish model shows that NAA15 and N-terminal acetylation defects cause heart disease | Independently confirms the causal link between pathogenic NAA15 variants and congenital heart disease | Burns lab @bostonchildrens.bsky.social www.biorxiv.org/content/10.1...
07.08.2025 12:25 — 👍 2 🔁 1 💬 0 📌 0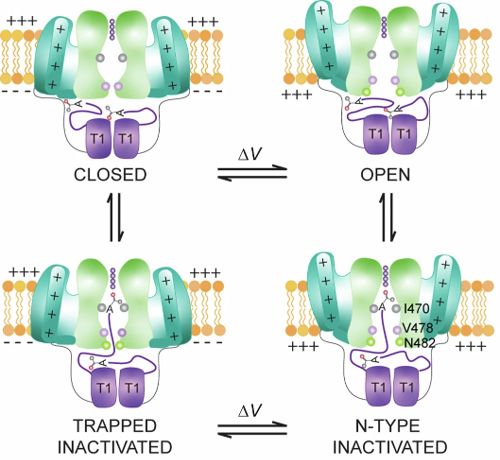
Mechanism for voltage-gated channel inactivation uncovered | N-terminal acetylation of the intracellular domain of the Shaker potassium channel is key for plugging the pore | By Kenton Swartz lab in @nature.com @ispt-proteinterm.bsky.social www.nature.com/articles/s41...
07.08.2025 08:11 — 👍 5 🔁 2 💬 0 📌 0
How to define degrons and E3 ligase specificities by peptide pulldowns | Methods in Enzymology | Kwon and Ji labs | #KCMF1 #Cysteine #Arg/N-degron @ispt-proteinterm.bsky.social www.sciencedirect.com/science/arti...
07.08.2025 07:11 — 👍 6 🔁 2 💬 0 📌 0
Using a specific antibody to detect proteins carrying N-terminal formylation | Methods in Enzymology | Hwang lab @ispt-proteinterm.bsky.social www.sciencedirect.com/science/arti...
07.08.2025 06:54 — 👍 2 🔁 1 💬 0 📌 0
Assessing protein N-terminal myristoylation. Detailed protocols by Ed Tate's group in Methods in Enzymology. #lipidation @ispt-proteinterm.bsky.social @imperialcollegeldn.bsky.social www.sciencedirect.com/science/arti...
06.08.2025 07:53 — 👍 4 🔁 1 💬 0 📌 0
Can we modulate plant oxygen sensing to enhance flood resilience?🌿🔧
www.biorxiv.org/content/10.1...
This is one of two preprints out this week from the @syno2xis.bsky.social and @emilyflashman.bsky.social labs, two angles on how PCOs shape plant flood resilience. Don’t miss the full story!

Expression and purification of ribosome associated factors: Methionine aminopeptidases and N-terminal acetyltransferases | Methods in Enzymology | Marius Klein and Irmgard Sinning @uniheidelberg.bsky.social @ispt-proteinterm.bsky.social www.sciencedirect.com/science/arti...
29.07.2025 07:46 — 👍 4 🔁 1 💬 0 📌 0
How to express, purify and assess the activity of the ATE1 arginyltransferase | Methods in Enzymology | Yi Zhang lab @ispt-proteinterm.bsky.social www.sciencedirect.com/science/arti...
29.07.2025 07:37 — 👍 3 🔁 2 💬 0 📌 0
Method to determine the identity and modification state of plastid precursor protein N-termini in the cytosol by combining protoplast protein import assays with targeted mass spectrometry | Methods in Enzymology | Baginsky lab. @ispt-proteinterm.bsky.social www.sciencedirect.com/science/arti...
29.07.2025 07:30 — 👍 2 🔁 1 💬 0 📌 0
Gid11 is a substrate receptor of the GID multisubunit E3 ligase during a metabolic switch. Specific degradation of proteins linked to glycolysis via their N-terminal threonine lacking N-terminal acetylation. #N-degron Khmelinskii lab @ispt-proteinterm.bsky.social
www.biorxiv.org/content/10.1...

Manipulating plant oxygen sensing through NCO substitution reveals trade-offs between growth and flooding tolerance. Perri et al. - another preprint from the Licausi and Flashman labs. @monicaperri1.bsky.social @emilyflashman.bsky.social @syno2xis.bsky.social www.biorxiv.org/content/10.1...
28.07.2025 08:40 — 👍 2 🔁 1 💬 0 📌 0
Engineering reduced activity in the oxygen-sensing Arabidopsis thaliana plant cysteine oxidase 4 enzyme results in improved flood resilience. By Dirr et al. Licausi and Flashman labs @syno2xis.bsky.social @emilyflashman.bsky.social www.biorxiv.org/content/10.1...
28.07.2025 08:35 — 👍 2 🔁 1 💬 0 📌 0Excited to announce our new methods paper in Methods in Enzymology!
We share our pipeline for building high-diversity DNA libraries & exploring protein stability in E. coli.
Free access until Sept 4: authors.elsevier.com/a/1lRkeHRzCX...
Updated, companion preprint: www.biorxiv.org/content/10.1...
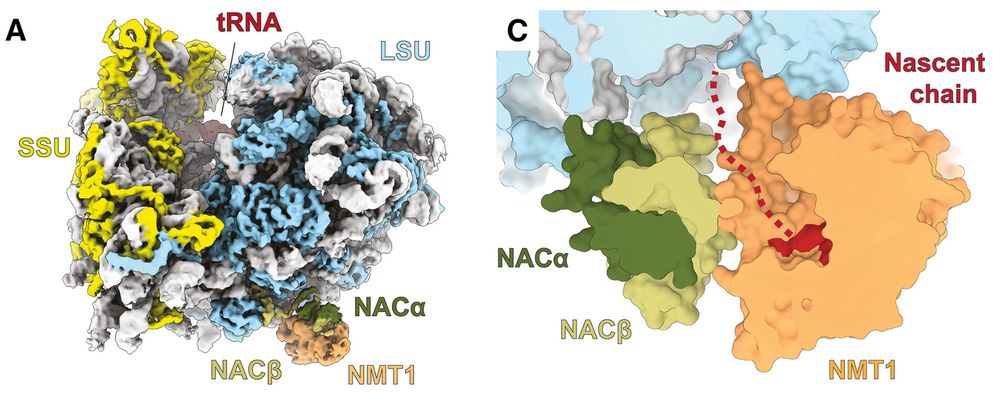
New paper out in Mol Cell, where we show how N-myristolytransferases lipidate proteins on the translating ribosome!
Continuing our successful collaboration with DeuerlingLab @uni-konstanz.de & Shu-ou Chan @caltech.edu
@snsf.ch @ethz.ch
www.cell.com/molecular-ce...