And one final comment - this work would not be possible without belief & funds for the idea to perform large-scale N-terminomics in patients (@mrinschen.bsky.social) and dozens of methods developed by/with colleagues: (S)HUNTER, MOFA, TPP-PISA and Human Kidney Organoids. Thanks for all this.
30.10.2025 21:19 — 👍 0 🔁 0 💬 0 📌 0

Summary scheme for our C3-LHF1 / SLE N-terminomics study; screening the proteolysis in SLE patients identified a cleavage in C3, which seems implicated in complement regulation (inhibiting CP & MBL pathways) and triggers IL6 signaling via JAK3/STAT3 - also increasing the levels of CXCL10.
To summarize: we identified the C3-LHF1 cleavage in a large-scale N-terminomics screen of a cross-sectional cohort which is increasingly present in SLE patients, inhibiting the CP & MBL pathways of the complement system while at the same time being a trigger for a IL6 response. Thanks for reading.
30.10.2025 21:11 — 👍 1 🔁 0 💬 1 📌 0

C3-LHF1, but not full-length human C3 triggers a IL6 reponse in HEK-Blue IL6 reporter cell lines. Partially a competition can be observed for IL6 in these reporter cell lines.
For further testing this interaction, we performed C3-LHF1 application to HEK-Blue IL6 reporter cell lines. We could observe a triggered IL6 response, albeit lower than IL6 agonist levels. The C3-LHF1 IL6 response can't be blocked by tocilizumab. There is even a little competition for IL6 & C3-LHF1.
30.10.2025 21:11 — 👍 0 🔁 0 💬 1 📌 0

C3-LHF1 pull-Down data from human kidneys identify IL6ST as one of the top interactors, especially as it is also one of the significantly stabilized proteins by C3-LHF1 addition in the TPP-PISA method.
Next, we screened putative interacting proteins for our rec. C3-LHF1 in human kidneys by classical pull-down assays and Thermal Proteome Profiling, PISA style): One of the top candidates is the IL6ST (aka gp130), a very important (co-)receptor for IL6 signalling.
30.10.2025 21:11 — 👍 0 🔁 0 💬 1 📌 0

We can observe the inhibitory effect of C3-LHF1 on the different pathways of the human complement system as analyzed by the SVAR Complement ELISA kits. C3-LHF1 strongly inhibits the MBL pathway and moderately inhibits the classical pathway - the alternative pathway seems not to be inhibited, but rather slightly activated at low concentrations.
We produced C3-LHF1 as a rec. protein & performed complement assays, revealing an inhibitory effect of C3-LHF1 on the classical and MBL pathway of the complement system. These assays were done with the SVAR Complement ELISA kits. Inhibiting complement can come in handy, but do we see other effects?
30.10.2025 21:11 — 👍 0 🔁 0 💬 1 📌 0

A circle plot combines obtained N-termini from the cross-sectional SLE cohort (in vivo), longitudinal perspective in remission (in vivo) and the recombinant complement protease assays (in vitro) - all pointing towards C3-LHF1 cleavage as a central element.
Combining all of this data into a fabulous circle plot clearly pointed towards an important role of the cleavage at pos. 1514, generating C3-LHF1. This became the focus of our future efforts. #LHF1 #CirclePlot
30.10.2025 21:11 — 👍 0 🔁 0 💬 1 📌 0

In-vitro N-terminomics assays identify substrate specificities and distinct cleavages for the proteases C1r, C1s and MASP-1/3. Especially in the C-terminal part of human C3, we could observe cleavages generated by MASP-1 and MASP-3, even with purified human C3 only, these cleavages were observed.
Knowing this, we wanted to know, if these cleavages could be generated by four proteases of the complement system: MASP-1/-3, C1r/s. We generated substrate profiles for these proteases & identified common substrates as well as intriguingly present cleavages at the C-terminus of C3, alike C3-LHF1
30.10.2025 21:03 — 👍 2 🔁 0 💬 1 📌 0

MOFA data analysis for the N-terminomics data is displaying significant correlations with clinical parameters and some factors feature also our already identified C3-LHF1 cleavage at pos. 1514. This cleavage is increased in SLE patients and among the top up-regulated determinants for the corresponding MOFA factor 5.
Generating mass spec data is trivial, but deriving conclusions is difficult, thus we applied the MOFA2 framework for analysis of our data. It yields significant correlations between some MOFA factors and clinical parameters and interestingly the presence of our C3-LHF1 cleavage in one of the factors
30.10.2025 21:03 — 👍 1 🔁 0 💬 1 📌 0

We can observe the Figure 2 C-E from the EMBOJ SLE paper and can see generally abundant processing of human C3 in plasma and especially enriched cleavage in the C3-LHF1 fragment in SLE patients compared to healthy controls.
Looking at proteolytis in human plasma, we observed a lot of cleavages in complement C3, with known cleavages for C3a, C3b, and C3(d)g - but also a cleavage at pos. 1514, which represented a quite intriguing lupus-enriched cleavage, thus we named it C3-LHF1 for complement "C3 Lupus Human Fragment 1"
30.10.2025 21:03 — 👍 0 🔁 0 💬 1 📌 0

A brief workflow for the SHUNTER protocol is depicted: firstly, denaturation of proteins including protease-generated N-terminus. Secondly, primary amines are protected by chemical modification via formaldehyde and subsequently digested with trypsin or similar. Newly generated peptide N-termini from the trypsin-digest are labeled with the aldehyde undecanal can be depleted in the next step by retention on C18 due to a higher hydrophobicity. Purified peptides from the flow-through contain the native protein N-termini from within the sample. Due to the higher incubation temperates, changed reagent and quicker turnaround, the workflow is much faster than previously.
With this new, improved SHUNTER protocol we can obtain better dimethylation, better N-termini purity and a higher identification rate in the end. Win-win for everyone!
Starting with an unbiased screening in human plasma, we needed to optimize our already published HUNTER method (doi.org/10.1074/mcp....) for high-throughput and the demands of handling a larger cohort with > 100 patients, optimizing efficiency. Stream-lined "label-free-like" N-terminomics. #SHUNTER
30.10.2025 11:09 — 👍 0 🔁 0 💬 1 📌 0
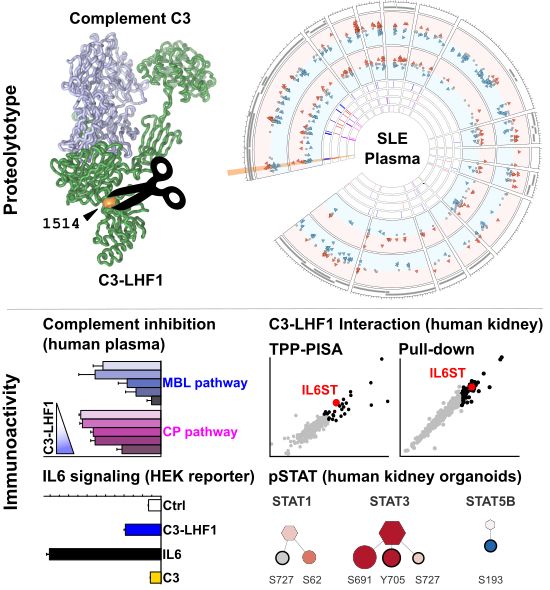
Investigation of the human plasma N-termini identifies the cleavage of the human complement C3 protein at position 1514 leads to generation of C3-LHF1. This C3-LHF1 significantly inhibits the classical and MBL pathways of the human complement system and interacts with the IL6ST/gp130 receptor, leading to IL6 signalling as evident from reporter cell line assays and (phospho-)proteomics screens.
After a looong journey, here the results of a deep investigation of human plasma proteolysis by MS: SLE N-terminomics paper with a complement C3 fragment at the core, EMBOJ: www.embopress.org/doi/full/10.... - highlights: a resource on proteolytically processed N-termini in human plasma #LHF1 #SLE🧪
30.10.2025 10:52 — 👍 1 🔁 1 💬 1 📌 0

See you at Palazzo dei Normanni in Palermo for the 2026 Protein Termini Conference | Co- and post-translational modifications | Proteolytic processing and degradation pathways | Microbes, plants, animals #ProteinTermini2026 #Proteostasis #ProteinModifications #PalazzoDeiNormanni #Palermo2026
26.10.2025 18:59 — 👍 6 🔁 7 💬 1 📌 1

Thalamic opioids from POMC satiety neurons switch on sugar appetite
High sugar–containing foods are readily consumed, even after meals and beyond fullness sensation (e.g., as desserts). Although reward-driven processing of palatable foods can promote overeating, the n...
Wer hat ihn, den berühmten Dessertmagen? 🙋🏼♀️
Ich fand diese Studie von vor ein paar Monaten super interessant, denn sie zeigt: Dieselben Nervenzellen, die uns nach dem Essen satt machen, setzen bei Zucker gezielt β‑Endorphin frei (= körpereigenes Opioid) das uns dazu treibt, weiter zu naschen (🐁)
03.07.2025 19:34 — 👍 60 🔁 7 💬 5 📌 3

Danish defense of Greenland sorted!
08.01.2025 16:28 — 👍 11 🔁 1 💬 1 📌 0

SCPro - Not single cell proteomics, at all, in any way, but still pretty cool!
Here, I fixed it for ya! Even though I can clearly see why I also wouldn't have recommended Ben Orsburn as a reviewer for this one, I do a...
I try super hard to be positive, but I seriously think we're at almost 9 out of 10 papers with "single cell proteomics" in the title don't feature the analysis of one actual cell in them anywhere. Case in point - super nice study. Obfuscating terminology. proteomicsnews.blogspot.com/2024/12/scpr...
03.12.2024 16:58 — 👍 18 🔁 5 💬 2 📌 1

I thought this poster of my facility turned out nice so I tight I’d share :)
22.11.2024 20:59 — 👍 31 🔁 3 💬 2 📌 0

Chemoproteomic mapping of the N-terminal cysteinome
The oxidation status of N-terminal cysteines directly dictates protein stability via arginylation and proteasomal degradation. However, only a handful of proteins have been shown to be regulated via t...
Interesting ChemRxiv preprint by the group of @tbpoulsen.bsky.social with first author @esbensv.bsky.social. They used OxSTEF probes to study proteins with N-terminal cysteines in the proteome and used the method to specifically investigate difference in normoxia and hypoxia. doi.org/10.26434/che...
21.11.2024 09:23 — 👍 16 🔁 4 💬 1 📌 0

If you are interested to learn about our research towards new targets for the antibiotics of the future, check out my eye-opener video. Thanks to the Leiden Institute of Chemistry, the Faculty of Science and the KNCV for making this possible.
www.eye-openers.nl/en/seethecas...
22.02.2024 16:33 — 👍 4 🔁 3 💬 1 📌 0

Eine Collage der neuen Produkte sowie einer Ansicht des kompletten Standes.
Alle Regalböden sind voll mit Kleidung, Playmobil und Schleichpferden in Tüten, Barbie-Puppen und -Zubehör und anderen Dingen.
Aufgefüllt und aufgeräumt!
Die #kirppudk 707 in #Tilst hat jetzt jede Menge Playmobil und Barbie, Schleichpferde, Mia&me, ...
26.11.2023 15:40 — 👍 3 🔁 1 💬 0 📌 0
Had there even been a real reason to use TEAB?
22.11.2023 11:08 — 👍 0 🔁 0 💬 1 📌 0

Single-use fast-food packaging hangs in the balance
France has already banned it — now industry is lobbying hard to ensure the EU doesn’t do the same.
Wer einen McDonald's im Ort hat, weiß, wie viel Müll dadurch überall herumliegt. Das wäre wirklich eine Verbesserung. Wären da nicht die Leute, die auch Pfandflaschen wegwerfen, weil zu viel Mühe ...
22.11.2023 09:26 — 👍 1 🔁 1 💬 0 📌 0

Happy to share the phenomenal PhD work of Dr. Amanda Lorentzian now out in its peer-reviewed from in @NatureComms. Amanda shows that targetable genome variants and proteome features are mostly stable from diagnosis to relapse in pediatric leukemia. 1/ www.nature.com/articles/s41...
22.11.2023 06:42 — 👍 4 🔁 1 💬 1 📌 0
 11.10.2023 01:09 — 👍 12 🔁 3 💬 1 📌 0
11.10.2023 01:09 — 👍 12 🔁 3 💬 1 📌 0
Inspired by bsky.app/profile/macc...
09.10.2023 07:46 — 👍 1 🔁 0 💬 0 📌 0

Google Scholar Wordcloud
There is for sure a better way to make the prime Blue Sky message more useful, but having a Google Scholar word cloud with your own publications might be a good one. So, let's see how the blue things evolve here #proteomics Kidneys in Arabidopsis would be a thing, I guess
09.10.2023 07:41 — 👍 0 🔁 0 💬 1 📌 0

Large-scale plasma proteomics comparisons through genetics and disease associations - www.nature.com/artic...
---
#proteomics #prot-paper
05.10.2023 10:00 — 👍 1 🔁 1 💬 0 📌 0
Interested in research ethics and all about #lupus #lupusnephritis | Running support group #Wales 🏴| Views my own | We need a cure for Lupus 🦋
Scientist, mother of two, love vacations and proteases 🇨🇴
Non-for-profit publication of research in molecular, cell & organismal biology - as part of @embopress.org
Fast-fair-transparent / Open Access / Open Science
Non-profit society of scientists devoted to understanding the molecular mechanisms and biological role of protein termini in plants, animals, fungi and bacteria. Conference every second year.
Website: https://proteintermini.org/
Das deutschsprachige @RealScientists.
Echte Wissenschaft von echten ForscherInnen, AutorInnen, KommunikatorInnen, KünstlerInnen... Diese Woche: @barteltlab.bsky.social
I hunt fake history and correct it, but also post amazing real history stuff. Rude, obscene, vulgar, racist people who can't act like grown-ups get blocked.
https://linktr.ee/fakehistoryhunter
Humorfacharbeiter und IrgendwasMitMedien
Biologist interested in membranes: signaling, trafficking, lipids, polarity, nanodomains, membrane contact sites, optogenetics, microscopy & plants. PI at @RDPlab @ENSdeLyon
@ERC_Research, alumn @salkinstitute, @EMBOFellow
Cancer #Proteomics Researcher | Mass Spec Enthusiast | Analytical Chemist | FFPE #Phosphoproteomics | She/her | My opinions are my own
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
She/her | Writer @MoGenUofT | PhD candidate @uoftmedicine @SinaiHealth @Gingraslab | I care about proteins and people, interested in mapping proteins, glycobiology, glycomics, mass spectrometry, and cancer #Proteomics #CancerResearch
Who: Staff Scientist
Where: IPMB, University of Zürich
What: Plants, peptides, receptors, PTMs, signaling
How: Molecular biology, biochemistry, proteomics
Why: Resiliant crops for the future
https://orcid.org/0000-0002-1805-8097
Scientist @Genentech love all small and automated devices, mass spectrometry, protein-protein interactions, cell-cell interactions, and how to make peptides/proteins flying into vacuum; View are my own
Computational Proteomics and Machine Learning fanatic.
Biological Mass Spectrometry enthusiast, trying to make sense of cellular hierarchies and heterogeneity
Lab news and updates for the Emmott lab, RNA virology & proteomics at the Centre for Proteome Research, University of Liverpool, UK
Lab website: emmottlab.org
Mentor, scientist & engineer. Having fun in @slavovlab.bsky.social and Parallel Squared Technology Institute @parallelsq.bsky.social with biology & single-cell proteomics.
https://nikolai.slavovlab.net
Professor of Genome Sciences University of Washington, Seattle. Interested in proteomics and mass spectrometry.























 11.10.2023 01:09 — 👍 12 🔁 3 💬 1 📌 0
11.10.2023 01:09 — 👍 12 🔁 3 💬 1 📌 0


