Congrats Guifeng! Great to see this out in its final form
09.09.2025 13:04 — 👍 0 🔁 0 💬 1 📌 0Joe Bowness
@joebowness.bsky.social
Postdoc @CRG.eu Velten lab 🥼🧬🩸 gene regulation & single-cell genomics | PhD from Brockdorff lab Oxford
@joebowness.bsky.social
Postdoc @CRG.eu Velten lab 🥼🧬🩸 gene regulation & single-cell genomics | PhD from Brockdorff lab Oxford
Congrats Guifeng! Great to see this out in its final form
09.09.2025 13:04 — 👍 0 🔁 0 💬 1 📌 0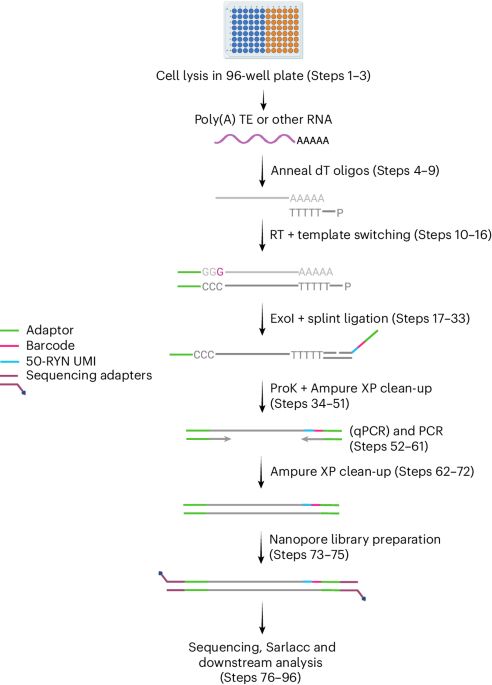
Very happy to share our protocols paper for CELLO-seq. This will make single cell long read RNA-seq more accessible and provides analysis guidelines. We hope this helps the #transposon #TEsky community and folks working on #singleCell isoform and allelic #gene expression. doi.org/10.1038/s415...
16.07.2025 16:55 — 👍 105 🔁 38 💬 8 📌 1I am very happy to have posted my first bioRxiv preprint. A long time in the making - and still adding a few final touches to it - but we're excited to finally have it out there in the wild:
www.biorxiv.org/content/10.1...
Read below for a few highlights...

Fresh preprint by @flavia-con.bsky.social from our lab uncovers how SMCHD1 finds & binds chromatin using live-cell single-molecule imaging🔬
She reveals how SMCHD1 dynamically engages chromatin, including the inactive X chromosome, to maintain gene silencing.
www.biorxiv.org/content/10.1...

Extremely proud of my sis for persevering through a huge number of roadblocks to get this article published! It's really important these stories are documented & get heard widely as stimuli to redress inequalities in systems of research doi.org/10.1186/s409... (she's now on a publishing 🔥 streak!)
20.06.2025 13:29 — 👍 2 🔁 0 💬 0 📌 0Monumental work! Huge congrats to all involved
21.05.2025 16:28 — 👍 5 🔁 1 💬 0 📌 0

It was such a pleasure to visit the beautiful cities of Bonn and Cologne, and have the opportunity to present my postdoc work at the @dzne.science. Thank you so much for hosting me @jsschrepping.bsky.social!
17.05.2025 13:25 — 👍 9 🔁 0 💬 1 📌 0Great to see the final form of this huge effort from the lab now published! Congrats Robert @juruehle.bsky.social @larsplus.bsky.social and all 🎉🎉
Read all about it this really cool work below ⬇️
Awesome! Congrats Iana (+co)!
08.05.2025 11:47 — 👍 3 🔁 0 💬 1 📌 0
Excited to share this story from the “Chromatin Dream Team” in the @arnausebe.bsky.social lab on chromatin evo across eukaryotes! 12 histone mark profiles from 12 species, including rhizarians, discobans, and cryptomonads. Read on to see what we found! www.biorxiv.org/content/10.1... 1/n
19.03.2025 12:02 — 👍 53 🔁 26 💬 1 📌 4Perfectly summarised! It was great to be part of such a fantastic event, and seamlessly cooperative organisation team. 😄
03.03.2025 08:56 — 👍 2 🔁 0 💬 0 📌 0Preprint alert. Excited to share our latest work. We demonstrate that the primary function of m6A on Xist is to promote RNA degradation. www.biorxiv.org/content/10.1...
10.01.2025 17:58 — 👍 4 🔁 1 💬 0 📌 1