Insightful seminar by Roderic Guigó: Figure 1 of the Craig Venter human genome paper (Science, 2001) was made by a Spanish PhD student who had no salary and earned his living serving tables in a Pizzeria.
27.02.2026 14:15 —
👍 5
🔁 1
💬 0
📌 1
Expansion cultures of mouse AML stem cells! Enabling large scale genetic screens in a system that's very close to "the real thing", with deep implications for drug discovery. Great work @alejofraticelli.bsky.social and @indrasingh.bsky.social
27.02.2026 14:12 —
👍 3
🔁 0
💬 1
📌 0
Early bird registration is closing Feb 28! We will have an AMAZING party!
17.02.2026 15:14 —
👍 2
🔁 3
💬 0
📌 0
Where are they from? They sing in bad Swahili or a very similar language, “I’m lighting fire, I’m lighting mama fire, and I’m bringing money”. Uganda military speaks (bad) Swahili, but does Museveni send soldiers to Ukraine?
11.01.2026 15:12 —
👍 3
🔁 0
💬 1
📌 0

Delighted to have our work on 🧬 resilience to 🩸cancer led by @g-agarwal.bsky.social & amazing collaborators, including @kharaslab.bsky.social, published in @science.org: www.science.org/doi/10.1126/... 🧵
02.01.2026 14:01 —
👍 56
🔁 15
💬 3
📌 1
There is one joker: Highly skilled technicians. Our two techs are why we can do projects that are ambitious, clear first author, no 100h weeks. Requires very strong commitment to a few projects by PI (more applicable to wetlab, and a + of 🇪🇸 is that there's no industry competing for the best people).
18.12.2025 10:26 —
👍 1
🔁 0
💬 1
📌 0
IMHO It’s near impossible to sustainably scale an academic research team quickly to 5-10 people. My take: Keep the bar for hiring high, keep money in the bank (allows you to scale up when things start working), and keep two full days per week free of Interruptions for hands on work.
15.12.2025 06:23 —
👍 6
🔁 1
💬 1
📌 0
@yusufroohani.bsky.social @rberrens.bsky.social @timcoorens.bsky.social @hsiuchuanlin.bsky.social @albarmeira.bsky.social @ferrannadeu.bsky.social @alejofraticelli.bsky.social @elimereu.bsky.social @simonhaas.bsky.social @leifludwig.bsky.social @stefaniegrosswendt.bsky.social
05.12.2025 10:35 —
👍 2
🔁 1
💬 0
📌 0

ISCO (Innovations in Single-Cell OMICS) will be back in beautiful Barcelona!
🗓️ 28th/29th of May 2026
📍Barcelona Biomedical Research Park @prbb.org (beachfront!)
Keynotes: @bartdeplancke.bsky.social and @bocklab.bsky.social
Submit your abstract and present your research!
www.isco-conference.eu
05.12.2025 10:32 —
👍 21
🔁 13
💬 2
📌 6

Yes, this is where you would work (the round building, not the boat)
We are looking for a postdoc to join our team! If you're interested in translating a cutting edge genomics technology (www.nature.com/articles/s41...) to real-life applications in hematology, this is for you. We offer a unique working environment ON THE BEACH: recruitment.crg.eu/content/jobs...
20.11.2025 09:36 —
👍 17
🔁 16
💬 0
📌 3
🗨️ Just published in Nature Biotechnology: Our CellWhisperer AI enables chat-based analysis of single-cell sequencing data. You can talk to your cells & figure out the biology without writing any computer code. Paper here: www.nature.com/articles/s41.... Annotated walkthrough in a thread below (1/11)
11.11.2025 12:52 —
👍 62
🔁 36
💬 2
📌 2

Are you looking for a PhD? Join us in Barcelona! You'll dive into a community of >100 PhD students from 30 countries exploring the frontiers of biology. You can also join an online workshop on 6 November (15:00 CET) to learn how to find the right lab for you.
More info: www.crg.eu/en/content/t...
23.10.2025 07:55 —
👍 25
🔁 28
💬 1
📌 8

Big, beautiful trees!!
SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life.
www.biorxiv.org/content/10.1...
14.10.2025 14:37 —
👍 46
🔁 15
💬 1
📌 0

Graphic with a blue background and light green dots. The text 'call for applications' appears in green, and 'Ausschreibung' in white. A white arrow on the right points diagonally upward.
Early-career researchers: want to run your own lab? 🌟Max Planck Research Groups offer 6+ years, up to €2.7M in funding, open-topic freedom, team support & tenure-track opportunities. Intrigued? 😃Apply by Oct 14, 2025! www.mpg.de/max-planck-r...
15.09.2025 09:26 —
👍 92
🔁 98
💬 0
📌 4
This was a lot of fun! Thanks Daylon and @arunsharmaphd.bsky.social for being great hosts!
18.09.2025 10:09 —
👍 5
🔁 2
💬 0
📌 0

Is the decline of reading making politics dumber?
As people read less they think less clearly, scholars fear
www.economist.com/culture/2025...
To counter this, here’s my 3 fav reads 📚 from this summer:
- The dream of the Celt by Mario Vargas Llosa
- Paradise by Abdulrazak Gurnah
- Dzhamilia by Tsjyngyz Ajtmatov
07.09.2025 05:13 —
👍 3
🔁 0
💬 0
📌 0
A really powerful, easy and affordable method for profiling cellular interactions in the immune system. Congrats @dom60.bsky.social , @simonhaas.bsky.social and everyone involved!
13.08.2025 12:11 —
👍 3
🔁 0
💬 0
📌 0

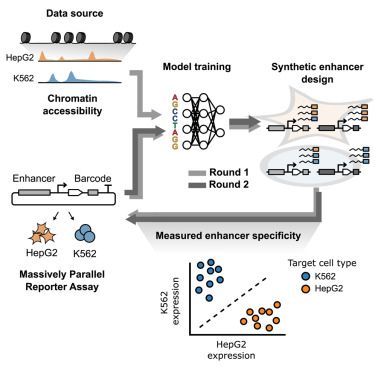
Beeswarm plot of the prediction error across different methods of double perturbations showing that all methods (scGPT, scFoundation, UCE, scBERT, Geneformer, GEARS, and CPA) perform worse than the additive baseline.

Line plot of the true positive rate against the false discovery proportion showing that none of the methods is better at finding non additive interactions than simply predicting no change.
Our paper benchmarking foundation models for perturbation effect prediction is finally published 🎉🥳🎉
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
04.08.2025 13:52 —
👍 126
🔁 57
💬 2
📌 6
Congrats @hoheyn.bsky.social , @lgmartelotto.bsky.social and team!
23.06.2025 07:13 —
👍 4
🔁 2
💬 0
📌 0

Scientists make breakthroughs on blood ageing and limb regrowth
Two new studies could help develop better therapies for rejuvenation and regeneration
🇬🇧: Finally, English-language coverage by Financial Times, a small London-based newspaper 😉. Thanks again @omarjam.bsky.social and Nahia (@irbbarcelona.org) for making all this possible! www.ft.com/content/a1f8...
31.05.2025 12:43 —
👍 1
🔁 0
💬 0
📌 0
😬: SER Catalunya, most-listened radio around here, interviewed me at 8:40 (instead of 8:50)... they reached me in the elevator, put me live on air and started asking in Catalan. Had to guess what they meant and improvise. The good thing, I was awake after this 😅 (can't upload audio to bsky - 7/n)
31.05.2025 12:43 —
👍 1
🔁 0
💬 1
📌 0