Of note, mosaic screens as performed by us or others can still be valuable tools, especially if the extent of heterogeneity is controlled by culturing conditions or quantified up front in absence of perturbations and taken into consideration by sufficient sampling and through biological replicates.
25.05.2025 11:00 — 👍 2 🔁 0 💬 1 📌 0
This platform is versatile (EBs are precursors for various organoids), scalable, and opens up new applications, such as studying genetic diseases with reduced penetrance or variable expressivity.
25.05.2025 11:00 — 👍 1 🔁 0 💬 1 📌 0
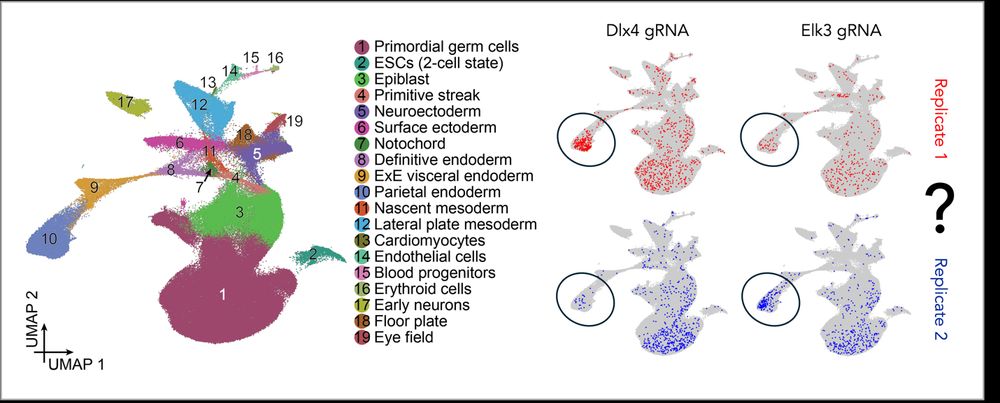
With organoid barcodes we can quantify baseline inter-EB heterogeneity and perturbation-specific changes in cell type composition. We can also apply replicate-based DE analyses, avoiding the high false positive rate caused by inter-EB heterogeneity in standard single cell DE.
25.05.2025 11:00 — 👍 1 🔁 0 💬 1 📌 0
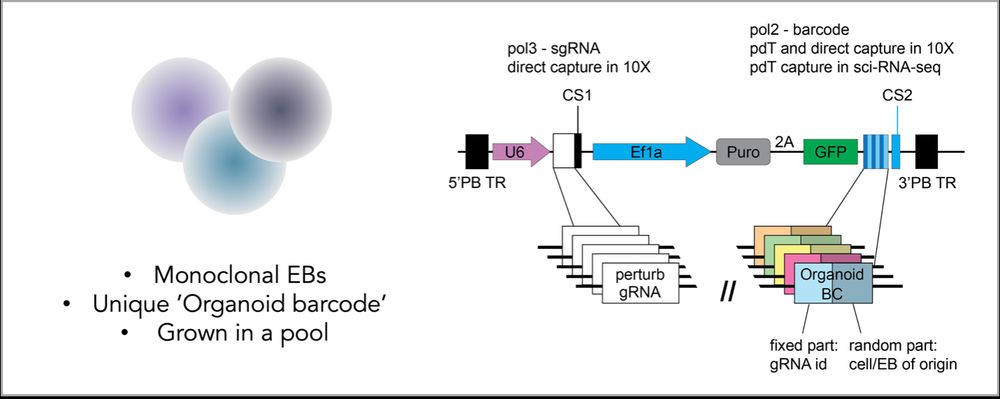
Our solution: monoclonal embryoids with an ‘organoid barcode’. This allows us to determine not only the transcriptome and perturbation identity for each cell but also the individual EB of origin - while still growing EBs in a pool, for scalability and shared culture conditions.
25.05.2025 11:00 — 👍 3 🔁 0 💬 1 📌 0
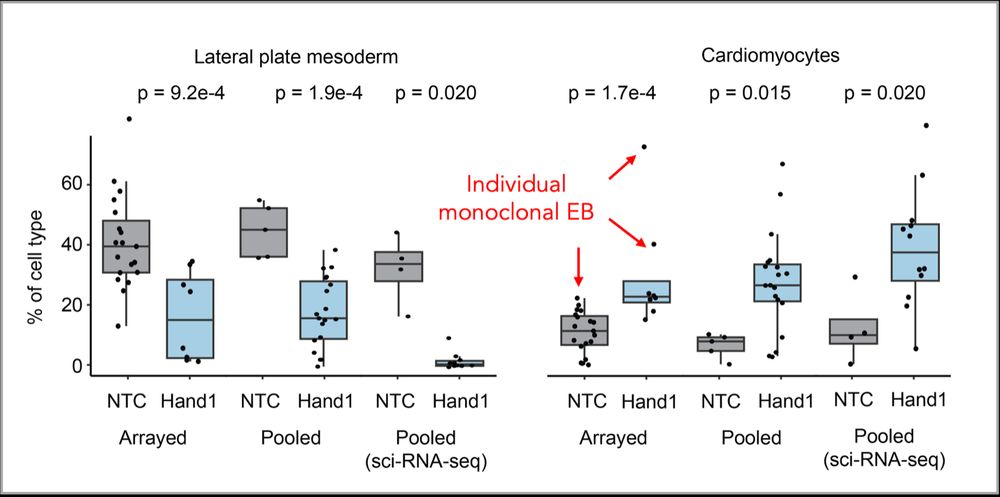
Many gRNAs seemed to have significant effects on cell type composition in mosaic EBs, but these were not reproducible across replicates: Stochastic clonal skewing of individual gRNAs in EBs, combined with inter-EB heterogeneity, overwhelms most perturbation effects and reduces statistical power.
25.05.2025 11:00 — 👍 2 🔁 0 💬 1 📌 0

Home - Kribelbauer Lab
USC Dornsife
How is it end of April already? Excited to share that the Kribelbauer Lab is up and running @USC/MCB. Grateful for my first two PhD students @m-finegan.bsky.social @christinagirgis.bsky.social. We are dev scalable, genome-int. tools to study TF & enhancer regulation. Website now live bit.ly/4jo8Tjg
29.04.2025 18:39 — 👍 21 🔁 6 💬 0 📌 0

a baby is eating a piece of food while sitting on the ground .
ALT: a baby is eating a piece of food while sitting on the ground .
Trying a slightly new model (for us at least) of #openscience. With @ronanchaligne.bsky.social and @karolisk.bsky.social, we pitted 10x Genomics Chromium GEM-X against Illumina PIPseq V head-to-head to understand these two platforms for single-cell genomics. Some thoughts off the bleeding edge: /1
14.03.2025 13:29 — 👍 59 🔁 18 💬 1 📌 3
Well said @dereklowe.bsky.social. Academia produces two vital products: (1) knowledge, and (2) human capital that knows how to use it. If we can’t train enough young creative US scientists to fulfill biopharma industry needs…
“I think they're being short-sighted, because fear does that to you.”
12.02.2025 19:41 — 👍 194 🔁 68 💬 3 📌 1

Stand Up And Be Counted
Biopharma companies and CEOs are keeping their heads down at their own peril. They should speak up about what’s happening to the NIH and other science agencies before it’s too late.
Silence gives consent. And no one should consent to this.
12.02.2025 16:45 — 👍 387 🔁 159 💬 17 📌 20
Nice work, congrats Sud!
03.02.2025 22:09 — 👍 1 🔁 0 💬 0 📌 0

CRISPR and Beyond 2025
Join us for the 6th CRISPR and Beyond Conference on 2-4 April 2025!🧬 A must-attend event for researchers in high-throughput screening and #GeneEditing, exploring emerging technologies and models. #CRISPR25
👉Programme and registration: bit.ly/3Amz34N
19.11.2024 14:40 — 👍 18 🔁 13 💬 2 📌 0
Senior Comp Bio Scientist @ www.gordian.bio |
Single-cell genomics and data viz enthusiast | Former Postdoc @ Harvard SCRB / Broad Institute w/ Buenrostro Lab
I'm an associate professor @UofUBiology
gagnonlab.org | fish stuff in and out of the tank
Assistant Professor of Neurobiology @ University of Utah investigating the neuroscience of social behavior and emotion.
UCLA > Caltech > U Utah
https://www.zelikowskylab.com
Gabilan Assistant Professor of Biological Sciences @ USC - Molecular and Computational Biology Section
Group Leader
The Genome Function Laboratory
The Francis Crick Institute, London
Professor at KTH, NY Genome Center, SciLifeLab, working on functional genomics and human genetics.
Aging, Systems Biology, Gene Therapy. CSO @GordianBio, doing mosaic therapeutic screens in vivo.
Also @NornGroup/Impetus Grants. Previously K99 Fellow @Buck Institute.
Banting Postdoc with @JShendure @uwgenome | Prev
@CIHR_IRSC UBC Neuro PhD | Genome engineering, Molecular Recording, neurodevelopment and its disorders | Mountains & skateboarding
statistical bioinformatics; canadian / swiss; #methodsmatter #rstats; open science advocate; grumpy towards prestige worshippers, excessive admin and bros; will sacrifice sleep for a sunrise.
https://robinsonlabuzh.github.io/
Associate Dean for data science, and Professor of Medicine and Human Genetics at the University of Chicago.
Scientist at UW Genome Sciences and the Seattle Hub for Synthetic Biology.
http://pinglay-lab.com/
synBio/genomics/soccer/heavy metal/food
Assistant Professor at UCSF | CZ Biohub – SF Investigator
https://diego-calderon-lab.github.io
https://pharmacy.ucsf.edu/diego-calderon
Head of mRNA Platform Design Data Science @Sanofi. R&D in mRNA therapeutics using ML/DL. Formerly @ UTAustin, MIT, MSFTResearch, UW, & Calico. Latest work: gScholar (https://tinyurl.com/3xdfksy6)
Assistant Professor, Rockefeller University
Incoming assistant professor at UC Irvine. Transcription factor control of cell state decisions using stem cell models of development. sskimlab.org
Senior Group Leader IMP / Adjunct Professor Medical University of Vienna. Fascinated by functional genetics (CRISPR, RNAi, degrons) and time-resolved omics towards understanding cancer biology and probing new therapeutic concepts.








