A screenshot of a dataset page on the cryoET data portal with the metadata panel opened and the View All Info button highlighted.
A paper on this is to follow, a variation was described here: www.nature.com/articles/s41...
An abridged version of the method can also be found directly on the cryoET portal. Click "View All Info" on the page of one of the datasets contained in this deposition.
28.01.2026 15:53 — 👍 1 🔁 0 💬 1 📌 0

Four representative slices of cryo-tomograms showing annotated organelles.
New on the CryoET Data Portal: ~27,000 tomograms of affinity-captured lysosomes from HEK293T cells across 4 physiological states. Includes raw data, AreTomo3 reconstructions & Membrain-Seg annotations. Openly available for reuse!
cryoetdataportal.czscience.com/depositions/...
27.01.2026 19:59 — 👍 72 🔁 20 💬 1 📌 3
This one was quite the journey! The paper describing the #ChlamyDataset is finally out and on the cover of Mol Cell!
This beautiful rendering made by co-author @jessheebner.bsky.social and Holly Peterson shows an instance of mitochondrial fission found in the dataset 😍
[Maybe long thread ahead]
09.01.2026 19:14 — 👍 90 🔁 35 💬 2 📌 10
Tired of knowing what you are looking for in your tomograms?
Why not try bluesky-less Frosina's new self-supervised algorithm for semantic segmentation and particle picking for #teamtomo (includes a new denoiser without the need for odd/even tomos)
Code is on GitHub if you want to try it out
19.11.2025 14:04 — 👍 52 🔁 11 💬 0 📌 0
CryoET Data Portal
Works with local OME-Zarr files, or as a bonus: stream sub-tilt stacks directly from the cryoET Data Portal: cryoetdataportal.czscience.com
Massive thanks to @danielji.bsky.social who did the heavy lifting implementing this over the summer! 🙌
17.11.2025 22:30 — 👍 5 🔁 0 💬 0 📌 0

GitHub - czimaginginstitute/zarr-particle-tools: Particle extraction and reconstruction from OME-Zarr tilt series.
Particle extraction and reconstruction from OME-Zarr tilt series. - czimaginginstitute/zarr-particle-tools
Hi #teamtomo! Want to use OME-Zarr for your tilt series but can't integrate it with your existing sub-tomogram averaging pipelines?
Try zarr-particle-tools - RELION-style extraction & reconstruction built for OME-Zarr-based workflows!
📦 pip install zarr-particle-tools
🔗 github.com/czimagingins...
17.11.2025 22:30 — 👍 16 🔁 7 💬 1 📌 0
Stunning cover to celebrate the incredible work from @loicaroyer.bsky.social's team.
Learn more about Ultrack, a powerful tool for fast, accurate and scalable cell tracking: biohub.org/life-science... 🧪
14.11.2025 19:23 — 👍 7 🔁 3 💬 0 📌 0

Biohub launches initiative combining frontier AI & frontier biology
The large-scale scientific effort combines frontier AI with frontier biology to accelerate discovery and solve disease.
EvolutionaryScale will be joining forces with Biohub to create the first large-scale scientific initiative combining frontier AI & frontier biology. Together, we're building world-class compute, AI research & state-of-the-art technologies to cure or prevent disease. bit.ly/4nOUGO4
06.11.2025 18:04 — 👍 3 🔁 1 💬 0 📌 0
Glushkova, Böhm, & Beck @maxplanck.de develop a publicly available computational method to measure the thickness of biological membranes in cryo-electron tomograms. Analysis of algae & human cells reveals systematic membrane thickness variations within & across organelles rupress.org/jcb/article/...
04.11.2025 17:30 — 👍 44 🔁 12 💬 1 📌 3

Figure 1

Figure 2

Figure 3

Figure 4
Lessons learned from a Kaggle challenge for particle picking in cryo-electron tomography [new]
Kaggle cryo-ET picking: Over-pick tol, aug, & annot key.
05.11.2025 10:48 — 👍 7 🔁 3 💬 0 📌 0

🎉 Our paper “MIFA: Metadata, Incentives, Formats and Accessibility guidelines to improve the reuse of AI datasets for bioimage analysis” is out in @natmethods.nature.com!
Community-driven standards to make bioimage data AI-ready & reusable.
👉 www.nature.com/articles/s41... #AI #Bioimaging #FAIRdata
15.09.2025 09:57 — 👍 16 🔁 8 💬 1 📌 1

GitHub - apeck12/denoiset: An implementation of Noise2Noise for cryoET data
An implementation of Noise2Noise for cryoET data. Contribute to apeck12/denoiset development by creating an account on GitHub.
New Title Alert: DenoisET- an implementation of the Noise2Noise algorithm specifically designed for cryoET data denoising.
Learn more here: buff.ly/8i8i87U
#SBGrid #SBGridSoftware #StructualBiology
14.08.2025 16:14 — 👍 7 🔁 2 💬 0 📌 0
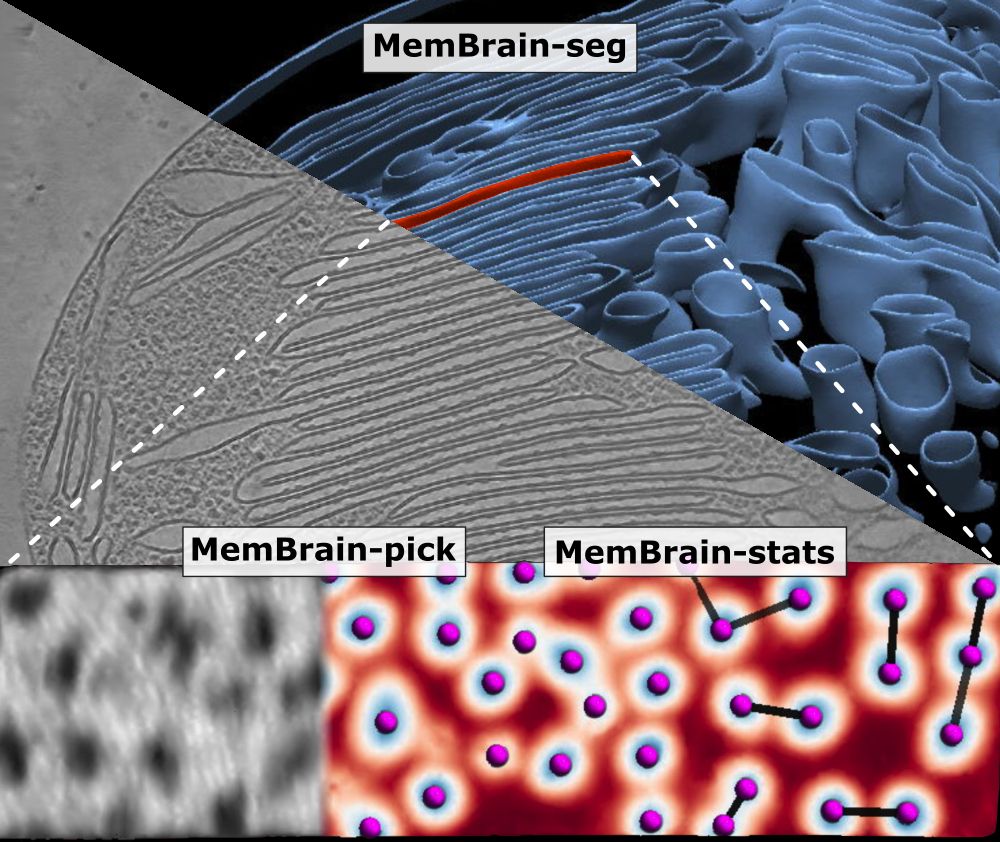
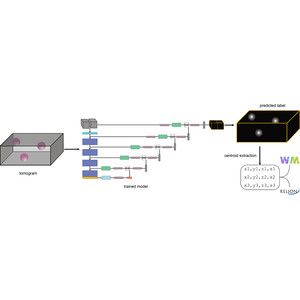
🦠🧠 MemBrain update! 🧠🦠
We’ve updated our preprint! It now covers the full MemBrain v2 pipeline for end-to-end membrane analysis in #CryoET: segmentation, particle picking, and spatial statistics.
🔗 Preprint: doi.org/10.1101/2024...
🔗 Code: github.com/CellArchLab/...
🧵(1/6) #TeamTomo
25.04.2025 07:28 — 👍 79 🔁 34 💬 2 📌 6
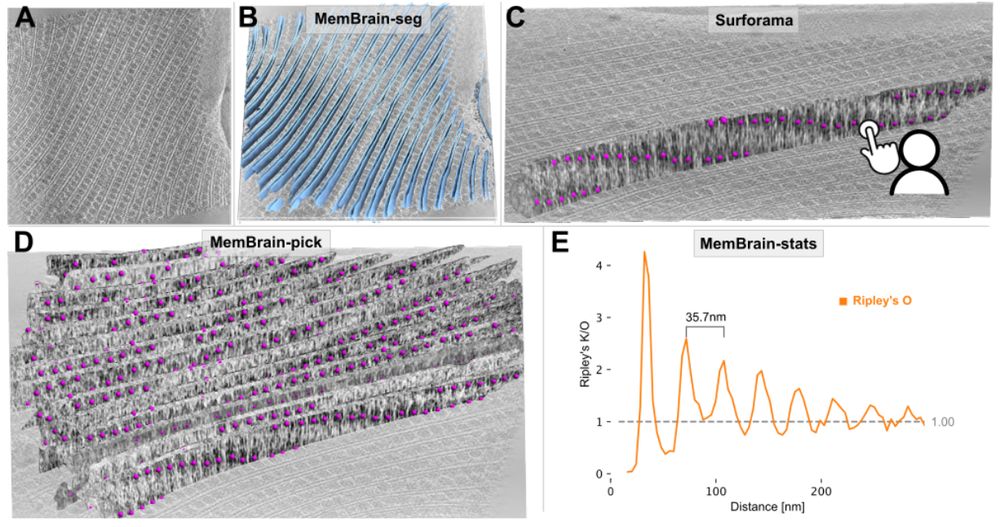
Figure 4. MemBrain v2 end-to-end workflow detects periodic phycobilisome organization. A: Raw tomogram slice of EMD-31244. B: Out-of-the-box MemBrain-seg segmentation (light blue). C: A single membrane instance can be visualized in Surforama and manually annotated with GT phycobilisome positions (magenta). D: MemBrain-pick localizes particles (trained with data from C) on all membranes in the tomogram. E: MemBrain-stats computes Ripley’s O statistic using the positions from D with a bin size of 5nm. The distance between peaks (35 nm) was measured to estimate chain unit spacings.
We have updated our #MemBrain v2 preprint with a lot more details about the MemBrain-pick and MemBrain-stats modules, as well as some application examples!
Stay tuned for the upcoming thread by lead author @lorenzlamm.bsky.social! 🧠🧵
#CryoET #TeamTomo
www.biorxiv.org/content/10.1...
23.04.2025 08:47 — 👍 69 🔁 20 💬 3 📌 1
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 — 👍 359 🔁 141 💬 12 📌 20
I'm super happy that our story is now published!
📖 www.science.org/doi/10.1126/...
But what changed compared to the original preprint?
Also, I feel i should post Movie 1 🎥, that inspired the cover. Back when I did the original bluesky thread, movies were not available.
21.03.2025 14:09 — 👍 215 🔁 68 💬 7 📌 9
The kaggle competition may be over, but we remain excited to see solutions from #TeamTomo!
If you're interested in checking whether your algorithm beats the kaggle winners' please don’t hesitate to give it a go and make a submission to the data portal!
06.03.2025 17:14 — 👍 2 🔁 1 💬 0 📌 0
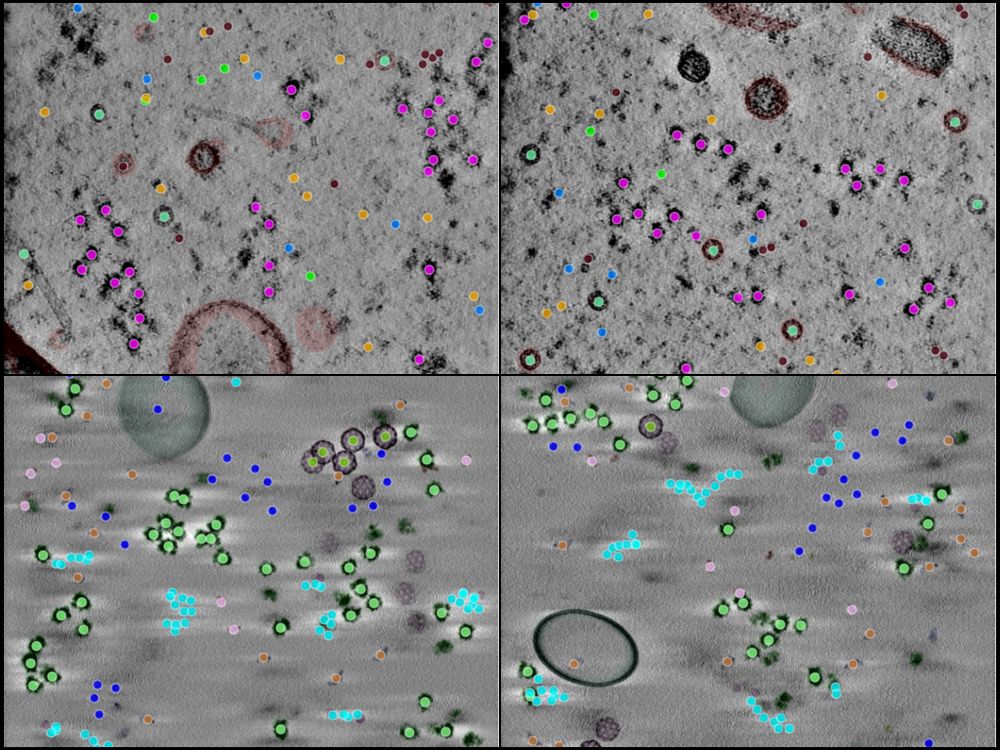
A 4-panel image showing tomographic slices and annotations from CZII's object detection challenge.
Now that the competition is closed, we're releasing all 492 runs, including raw data, tomograms and ground truth annotations as a benchmark dataset for the community. Find the datasets here: cryoetdataportal.czscience.com/depositions/...
06.03.2025 17:14 — 👍 5 🔁 1 💬 1 📌 0
Results from CZII's #cryoET object detection challenge on kaggle are in!
Participation exceeded our expectations with 931 Teams from 76 countries competing, generating over 27,000 submissions. For an overview check out our post-competition page: cryoetdataportal.czscience.com/competition
06.03.2025 17:14 — 👍 13 🔁 3 💬 1 📌 1
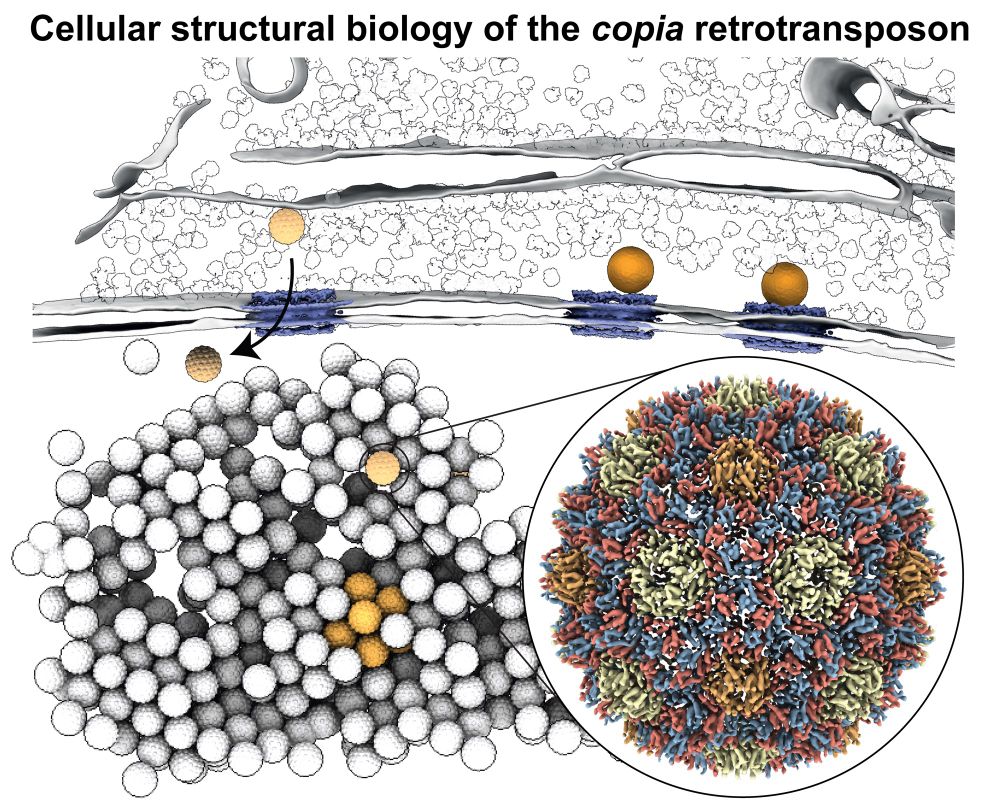
Happy to share our manuscript on the in situ visualization of the copia retrotransposon in its final form today published in @cellcellpress.bsky.social www.cell.com/cell/fulltex.... What’s new?
05.03.2025 16:02 — 👍 195 🔁 79 💬 12 📌 10
Exciting news! Are you planning to eventually release these beautiful segmentations as well?
28.02.2025 21:26 — 👍 2 🔁 0 💬 1 📌 0
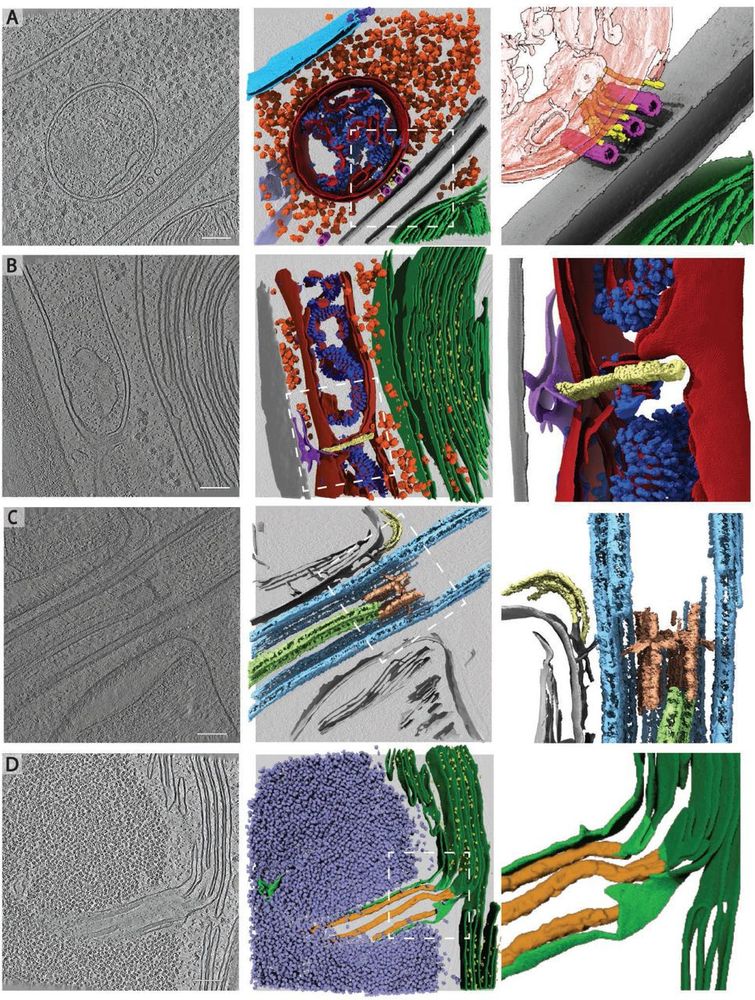
Rare cellular events observed in the cryo-ET dataset. Slices through tomograms (left) with corresponding 3D segmentation (middle and right). Right panels show enlarged views of the boxed regions in the middle panels. A) Mitochondrion at the plasma membrane, tethered to microtubules by thin filaments of unknown identity. Segmented classes: nuclear envelope (light blue), nuclear pore complex (dark blue), 80S ribosomes (orange), mitochondrial membranes (dark red), ATP synthase (royal blue), plasma membranes (grey), microtubules (pink), filaments (yellow), endoplasmic reticulum (purple). Zoomed-in panel shows thin filaments running between microtubules and the mitochondrion at the plasma membrane. Note that this tomogram contains two closely appressed cells, and thus, two plasma membranes with cell walls between. B) Mitochondrial fission event with ER membrane interactions. Segmented classes: mitochondrial membranes (dark red), ATP synthase (royal blue), 80S ribosomes (orange), thylakoid membranes (green), PSII (bright yellow), cell wall (grey), ER (purple), fission site containing filamentous structures perpendicular to the mitochondrial long axis (pale yellow). C) Ciliary transition zone between basal body and axoneme, including assembling IFT train and stellate structure77. Segmented classes: microtubule doublets (light blue), central microtubule pair (light green), stellate structure (pale red), IFT train (yellow). D) Pyrenoid tubule extending from the thylakoids into the phase-separated Rubisco matrix of the pyrenoid. Minitubules originate from thylakoid membranes82. Segmented classes: thylakoid membranes (dark green), PSII (yellow), pyrenoid tubule (lime green), Rubisco (lavender blue), minitubules (orange). Scale bars in A-D: 100 nm. Related to Fig. 2C-H.
To all #TeamTomo #CryoET and #Chlamydomonas aficionados: we have updated EMPIAR-11830 with a bug fix concerning some cryo-CARE denoised tomos as well as additional files and metadata! 🎉👩🏽💻
www.ebi.ac.uk/empiar/EMPIA...
A little thread about what's new... 🧵 1/n
28.02.2025 16:35 — 👍 64 🔁 22 💬 2 📌 1
Check out the press release about our recent Nature Methods publication on Segment Anything for Microscopy!
24.02.2025 17:27 — 👍 25 🔁 2 💬 1 📌 0

Synaptic vesicles segmented with SynapseNet.
Our first deposition of synaptic vesicles segmentations is now in the Cryo ET Portal! We segmented vesicles in over 50 tomograms to enable analysis of membrane proteins and more.
cryoetdataportal.czscience.com/depositions/...
21.02.2025 06:57 — 👍 50 🔁 10 💬 2 📌 1
FakET: Simulating cryo-electron tomograms with neural style transfer pubmed.ncbi.nlm.nih.gov/39947174/ #cryoEM
14.02.2025 20:33 — 👍 6 🔁 3 💬 0 📌 0

Excited to share that Eugene Khvedchenia and I secured 🏆 1st place out of 950 teams in the CryoET Object Identification Competition hosted by the Chan Zuckerberg Institute for Advanced Biological Imaging (CZII) on Kaggle! www.kaggle.com/competitions...
07.02.2025 12:48 — 👍 13 🔁 1 💬 1 📌 0
Hey #teamtomo, if you often find yourself using the excellent membrain-seg from @lorenzlamm.bsky.social et al., you might find my napari plugin "napari-segselect" useful. Let's say your tomogram contains the edges of two bacterial cells, each with a membrane and cell wall:
06.02.2025 11:02 — 👍 37 🔁 12 💬 3 📌 1
IGBMC near Strasbourg
DNA and chromatin in situ
https://x.com/EltsovMikhail
Group Leader (Organelle Biology) @ Biohub, Redwood City, CA
scientist in #cryoem and professor @uni-wuerzburg.de
https://orcid.org/0000-0003-3471-4637
Plants, Pathogens and Procrastination
Emmy Noether Fellow at ZMBP
Interested in protein structure and evolution. Somehow what they call a "PI" now, not sure how that happened. Hiring!
Solver of GTPases molecular puzzles | Membranes | CRISPR | STED super-res |Accidental immunologist | Academic mother of 2 cheeky monkeys | Assistant prof at FU Berlin. | Head of the Nanoscale Membrane Dynamics Lab
Co-Creating Ireland's Public Involvement in Open Research Roadmap
ENGAGED is building a national roadmap to shape public involvement in open research in Ireland. We believe that research can and does play an important role in tackling societal challenges.
The Electron Microscopy Data Bank @ EMBL-EBI. Core wwPDB consortium partner. Posts are made by our data release pipeline :) and ad-hoc by team members. Views and opinions are our own.
EMDB Team Leader - EMBL-EBI. wwPDB Principal Investigator. Fathering, mixology & DIY enthusiast. #CryoEM #CryoET #firstgen. All opinions are my own.
Cell adhesion, cytoskeletal regulation, Wnt signaling & wherever science leads us + wildflowers & my own idiosyncratic views. First gen college grad
Diversity, equity & Inclusion are core American Values
https://peiferlab.web.unc.edu
middle child, creator of anywidget.dev
building @marimo.io; prev @harvard.edu
Research Scientist in Loic Royer's team at Biohub | 🇧🇷
AI/ML Research Scientist @ Bi[o]hub
Combining frontier AI & frontier biology to help scientists cure or prevent disease
Journal of Cell Biology publishes peer-reviewed research on all aspects of cellular structure and function. Published by Rockefeller University Press @rupress.org
🌐 https://rupress.org/jcb
Neurologist and Alzheimer disease scientist at Brigham and Women’s Hospital. My lab studies beta-amyloid aggregates as they occur in human brain. Also TDP-43 biomarkers. Associate PD for research at MGB neurology residency program. sternlab.bwh.harvard.edu
https://isb.med.upenn.edu/