Excited to see a home for MCP servers in bioinformatics emerge! The community needs a central hub for developing MCP servers for key bioinfo tools. At @scverse.bsky.social we're contributing to this effort - can't wait to share what we're building
06.11.2025 19:28 — 👍 6 🔁 3 💬 0 📌 0
I'm recruiting a postdoc for my group (based in beautiful Eugene, OR). Please get in touch if you're interested, esp if you'd like to chat at #ASHG25!
15.10.2025 12:52 — 👍 39 🔁 42 💬 0 📌 1
👉 and make sure not to miss the one-and-only Romain Lopez's talk about PhenoBridge, our joint work on ML approaches to integrate perturb-seq and genetic association data (Wed 10.45am)
Come find me or just reach out if you'd like to chat! [2/2]
13.10.2025 23:19 — 👍 2 🔁 1 💬 0 📌 0
En route to Boston for #ASHG25 #ASHG2025!
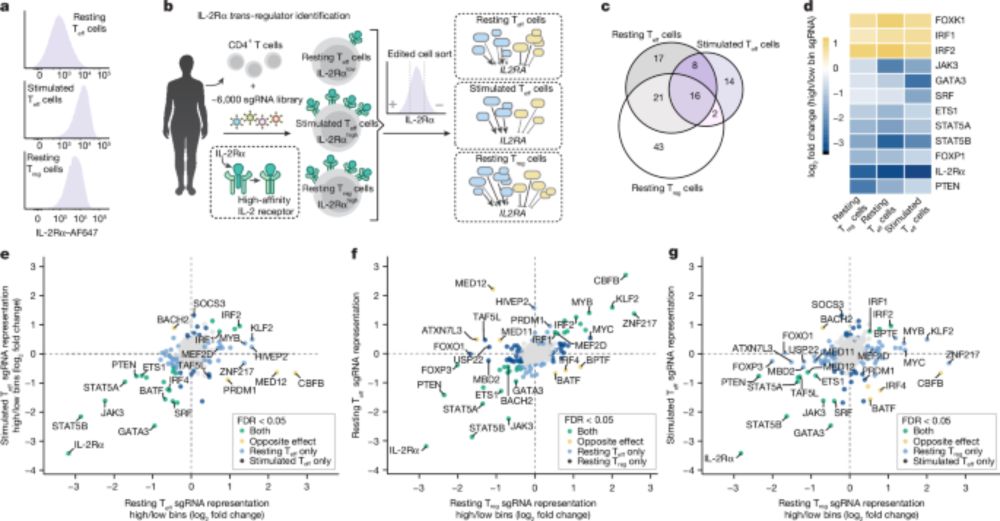
👉 I'll be presenting a poster on our new work on genome-wide perturb-seq screens in primary human T cells (5049W, Wed 2.30pm)
👉 you can hear me talk about it at the Industry Education session presented by Ultima Genomics (Thu 3pm) [1/2]
13.10.2025 23:19 — 👍 7 🔁 2 💬 1 📌 0

Last call, scverse community!
🐦 Early-bird closes October 17 for the scverse Conference 2025.
Single-cell, open science, and the people behind it — all in one place.
📝 Register here: www.eventbrite.com/e/scverse-co...
2 days of talks + 1 day of workshops
ℹ️ Info: scverse.org/conference2025
10.10.2025 02:48 — 👍 2 🔁 1 💬 0 📌 1
🚀 NEW PANEL ANNOUNCEMENT: Agentic Workflows in Bioinformatics at scverse 2025! 🚀
We're thrilled to introduce an exciting panel discussion at the upcoming scverse Conference 2025!
More information in 🧵
#scverse2025 #Bioinformatics #AgenticAI #Conference
03.10.2025 15:43 — 👍 7 🔁 4 💬 1 📌 2

🧬 Tahoe Workshop at scverse conference 2025 🧬
Disease Biology and Therapeutics in the Age of Frontier Datasets
🧵
@tahoetherapeutics.bsky.social
#scverse2025 #ComputationalBiology #DrugDiscovery #TahoeTherapeutics #SingleCell #PerturbationBiology
02.09.2025 16:03 — 👍 3 🔁 2 💬 1 📌 0

🎉scverse conference 2025 Call for Abstracts DEADLINE EXTENDED! 🎉
We're excited to announce that the deadline to submit abstracts for the scverse Conference 2025 has been extended to September 15, 2025!
🧵
#scverse #scverse2025 #SingleCell #Conference
01.09.2025 16:17 — 👍 5 🔁 5 💬 1 📌 0

Meet the keynote speakers for the 2025 scverse conference!
Elham Azizi, Associate Professor of Cancer Data Research and Biomedical Engineering at Columbia University
🧵
@elhamazizi.bsky.social
@columbiauniversity.bsky.social
#scverse #scverse2025 #machinelearning #genomics #cancerimmunology
28.08.2025 15:04 — 👍 13 🔁 5 💬 1 📌 0

🧫 Meet the keynote speakers for the 2025 scverse conference!
Erika Alden DeBenedictis, Co-founder of The Align Foundation & CEO of Pioneer Labs
@erika-alden.bsky.social
@alignbio.bsky.social
@pioneerlabs.bsky.social
#scverse #scverse2025 #StructuralBiology #Biotech #Stanford
26.08.2025 14:36 — 👍 8 🔁 6 💬 1 📌 0
We're committed to support as many attendees as possible join us at #scverse2025 - feel free to reach out if you have questions!
25.08.2025 17:17 — 👍 4 🔁 3 💬 0 📌 0

🧠 Meet the keynote speakers for the 2025 scverse conference!
Panos Roussos, Professor at Icahn School of Medicine at Mount Sinai
🧵
@panosroussos.bsky.social
#scverse #scverse2025 #SingleCell #SpatialTranscriptomics #Conference #Keynote
19.08.2025 15:37 — 👍 3 🔁 2 💬 1 📌 1
🎉 scverse conference 2025 Registration & Call for Abstracts NOW OPEN! 🎉
We're excited to announce that registration and the call for abstracts are officially open for the scverse Conference 2025!
Details in thread!
🧵 1/3
02.08.2025 15:00 — 👍 24 🔁 13 💬 1 📌 7
I have an opportunity to hire a staff scientist for my lab. Looking for someone with outstanding skillset in ML/statistics, genomics applications; interest in mentoring, strong publication record, PD experience required.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
01.06.2025 15:33 — 👍 83 🔁 108 💬 3 📌 3
It's happening! 🙌
12.05.2025 23:24 — 👍 6 🔁 0 💬 0 📌 0

Release notes
Version 1.11: 1.11.0 2025-02-14: Release candidates: rc2 2025-01-24, rc1 2024-12-20. Features: rc1 sample() supports both upsampling and downsampling of observations and variables. subsample() is n...
🎉 Scanpy 1.11.0 is out! 🎉 just after reaching 2000 stars on GitHub!
- sc.pp.sample replaces subsample with many new features
- Sparse Dask support pca
- session-info2 package for more reproducible notebooks
See the release notes:
14.02.2025 12:08 — 👍 49 🔁 19 💬 1 📌 1

Donate to Half Iron(wo)man for Meningitis Research, organized by Sophie Belman
I will be racing my first Half Ironman triathlon in Zurich this June. It includ… Sophie Belman needs your support for Half Iron(wo)man for Meningitis Research
I’m racing a Half Ironman triathlon this June. 2km swim, 90km cycle, 20km run and raising money for the Meningitis Research Foundation.
Meningitis has ~2.5 million cases and 250,000 deaths annually predominantly among children. Any donation would be great!
gofund.me/a958252f
09.02.2025 19:29 — 👍 4 🔁 2 💬 1 📌 0


Come work at the European Molecular Biology Laboratory in the beautiful science & university city Heidelberg as a Research Software Engineer on R/Bioconductor tools for biological data science and AI !
embl.wd103.myworkdayjobs.com/EMBL/job/Hei...
31.01.2025 16:03 — 👍 34 🔁 17 💬 2 📌 1

🚨🚨 MEGA JOB ALERT 🚨🚨
Independent Group Leader Positions in Computational Biology @humantechnopole.bsky.social!
Are you ready to start your own lab? Do you know someone who is? Repost this + share with everyone who might want to know about it. Thanks!!! 🙏
More details below... check it out! 🧵 1/3
29.01.2025 10:39 — 👍 57 🔁 63 💬 1 📌 2
I posted a couple days ago about our new paper on building causal graphs from genetic associations + Perturb-seq.
Here I want to expand on the value of using DIRECTIONAL information contained in LoF burden tests.🧵
[work led by @minetoota.bsky.social ]
bsky.app/profile/jkpr...
27.01.2025 19:18 — 👍 61 🔁 14 💬 1 📌 1
@minetoota.bsky.social set the groundwork for many ongoing projects in @jkpritch.bsky.social and Marson lab. Great to see this out!
26.01.2025 19:29 — 👍 10 🔁 3 💬 0 📌 0
Very excited about this new work from our lab! Explainer thread coming soon
@minetoota.bsky.social
24.01.2025 16:45 — 👍 62 🔁 28 💬 1 📌 0
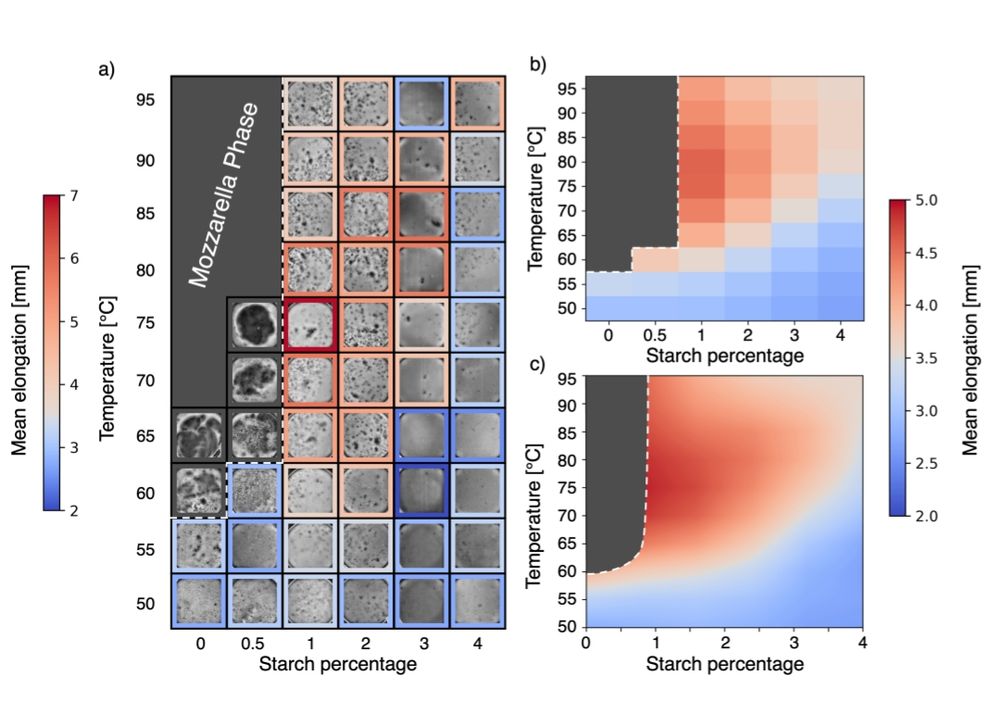
1/n Some time ago my colleague, excellent cook, and friend Ivan told me: "Cacio e pepe is the recipe that I screw up more often. Let's make a project studying systematically the physics of that sauce".
Prepare to get cheesy, I'm glad to share the Cacio e paper preprint:
arxiv.org/abs/2501.00536
04.01.2025 09:33 — 👍 498 🔁 167 💬 25 📌 68
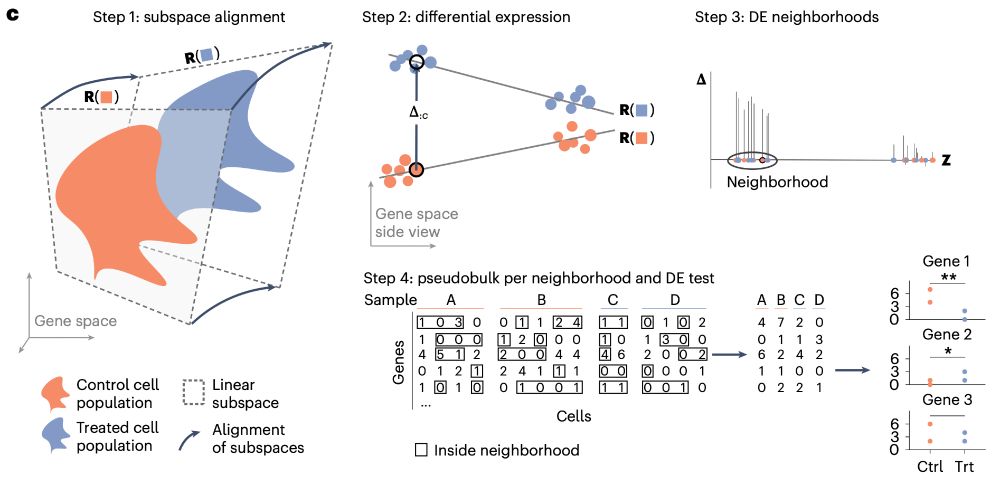
Overview of the LEMUR steps: (1) subspace alignment, (2) differential expression, (3) DE neighborhoods, (4) pseudobulking.
After 4y in the making, I am super excited that my main PhD project is published 🎉🥳🎉🎉🥳
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
03.01.2025 19:25 — 👍 155 🔁 35 💬 8 📌 3
Specificity, length, and luck: How genes are prioritized by rare and common variant association studies https://www.biorxiv.org/content/10.1101/2024.12.12.628073v1
16.12.2024 10:33 — 👍 46 🔁 24 💬 0 📌 1

# Define model with interaction term
model = MyModel(data, "~ treatment * timepoint")
# compare timepoints
contrast = model.cond(timepoint="on_treatment") - model.cond(timepoint="baseline")
# compare timepoints within drugA only
contrast = (
model.cond(treatment="drugA", timepoint="on_treatment") -
model.cond(treatment="drugA", timepoint="baseline")
)
# compare interaction of timepoint with treatment
# (= difference of changes between both treatments)
contrast = (
mod.cond(treatment="drugB", timepoint="on_treatment")
- mod.cond(treatment="drugB", timepoint="baseline")
) - (
mod.cond(treatment="drugA", timepoint="on_treatment")
- mod.cond(treatment="drugA", timepoint="baseline")
)
Formulaic is the go-to way to specify design formulas in Python, e.g. ~treatment + timepoint.
To compare sth, one needs to specify a contrast, e.g "on treatment vs baseline".
To make this easier, we developed "formulaic-contrasts":
formulaic-contrasts.readthedocs.io/en/latest/
07.12.2024 19:05 — 👍 18 🔁 6 💬 1 📌 1

Seeing all my old friends on here again
16.11.2024 18:42 — 👍 12071 🔁 1077 💬 92 📌 55
Journalist covering biotechnology for MIT Technology Review. Scoops about new methods in genetics and cell biology. The "playing God" beat. Gene editing, gene therapy, ancient DNA, synthetic embryos, cloning, etc.
PhD student at the Yosef Lab at the Weizmann Institute of Science | Computational Biologist
Assistant Professor @uarizona; macro-evolution, data science, and some ecology; Lab website: https://datadiversitylab.github.io/; Blog: https://ghost.cromanpa.synology.me/
Postdoc @broadinstitute.org @harvard.edu #MGH - Immunogenomics - PhD in Genomics🧬 @cam.ac.uk @sangerinstitute.bsky.social🔬 #LCG-UNAM
Assistant Professor, BME, Columbia University
PhD. Candidate | Seoul National University ‘19~| University of Pennsylvania ~’18 | Berkshire School ~’12 | Bioinformatics | Single Cell Analysis | ASC Organoids
Studying environmental impacts of computing and promoting Environmentally Sustainable Science 🌱
Researcher @CambridgeUni | Green Algorithms | Green DiSC | Fellow Software Sustainability Institute | he/him | 🐕 picture
https://www.lannelongue-group.org
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Assistant Professor in Bioimaging and Data Analysis and Director of the BioVisionCenter at UZH; Research Group Leader at EMBL-EBI. Loves image analysis, mountains, and birds.
Incoming postdoc at Genentech | PhD from Stanford Biomedical Data Science.
Assistant Professor of Bioengineering, Berkeley. Living in an RNA world. lareaulab.org
Assistant Professor, Cold Spring Harbor Laboratory
Systems immunology to understand thymus physiology and T cell development
@RingAScientist, @SkypeScientist
Statistical genetics - genetic architecture and algorithms - Assistant Professor at Harvard DBMI
Building AI4Science Foundation Models and AI Agents for Biomedicine, Accelerating the Engineering of Life, be in on Earth, on Mars, and Beyond!
Assistant Professor in the Section of Computational Biology, Division of Oncology, Dept. of Medicine at WashU
scverse Core Member & Community Manager
Evans MDS Young Investigator
jfoltzlab.org
Postdoctoral Fellow at Marioni Lab, Genentech | Aging & bioinformatics
Born in Portugal 🇵🇹 living in California 🐻
Stanford Genetics PhD Candidate
MIT Physics 2020
I also ride bikes
The Institute for Human Genetics (IHG) is the hub for genetics/genomics research, technologies, industry partnerships, training, and education at UCSF.