
Legit work in progress photo…
www.imperial.ac.uk/news/271753/...
@tomstantonmicro.bsky.social
Microbiologist. Lead developer of #Kaptive + bonafide #Klebsiella nerd. Post-doc in the Wyres Lab @AlfredMonash_ID.

Legit work in progress photo…
www.imperial.ac.uk/news/271753/...
Hard to believe it’s already that time of year!
Our kick-off meeting for the Klebsiella Seminar Series just wrapped up. Stay tuned for more!
@lauraamike.bsky.social @olayarendueles.bsky.social @caityholmes.bsky.social @tomstantonmicro.bsky.social @juanvalenciabacca.bsky.social WenWen Low & Jay V.
The @klebnet.bsky.social team are pleased to share slides from our “Klebsiella pneumoniae Genomic Epidemiology & Antimicrobial Resistance” lecture series!
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...

Output from Kaptive analysis shown in a BIGSdb isolate record. Tabular results are shown followed by a graphical representation of the genes within the locus analysed.
BIGSdb v1.51.4 has been released. This adds a new #Kaptive plugin for surface polysaccharide typing of Acinetobacter baumannii and Klebsiella. github.com/kjolley/BIGS... for details. Kaptive is developed by @tomstantonmicro.bsky.social, @kelwyres.bsky.social , @katholt.bsky.social and colleagues.
18.07.2025 07:57 — 👍 10 🔁 4 💬 1 📌 2Our new #klebsiella O type nomenclature, codesigned with Chris Whitfield, is now live in @pathogenwatch.bsky.social!
Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
We are pleased to launch the KlebNET Genomic Epidemiology Consortium!
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
Always a pleasure to organize this with @caityholmes.bsky.social @lauraamike.bsky.social Jay Vornhagen, Wen wen low, & this year joining us @tomstantonmicro.bsky.social & Juan Valencia.
01.04.2025 08:30 — 👍 2 🔁 1 💬 0 📌 0Janitor saves the day again!
www.rdocumentation.org/packages/jan...

K. variicola can be misID as K. pneumoniae. Our #OpenAccess paper reports an NICU #Outbreak
📌The rapid detection of a neonatal unit outbreak of a wild-type Klebsiella variicola using decentralized Oxford Nanopore sequencing
doi.org/10.1186/s137...
@nanoporetech.com
🖥️🧬💻
#AcademicSky
#Microsky
🧪🧫🦠

New pre print! We establish an ex vivo blood vessel model to investigate the effect of infection on vascular physiology in real time. We show Klebsiella inhibits vasodilation in a T6SS-dependent manner by targeting eNOs. Superb work by @safimicro.bsky.social www.biorxiv.org/content/10.1...
06.02.2025 09:15 — 👍 36 🔁 18 💬 0 📌 1Lastly, we'd like to thank YOU, the Kaptive community, for guiding development, spotting bugs and collaborating with us!
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
Kaptive 3 is now integrated within Kaptive-Web (kaptive-web.erc.monash.edu), PathogenWatch (pathogen.watch), the new Kleborate 3 framework (github.com/klebgenomics...) and Bactopia (bactopia.github.io/latest/).
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
We know the command-line can be tricky, so we made the CLI much friendlier 🧑💻
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/

Kaptive 3 is also much (much) faster than Kaptive 2, taking ~1 second per assembly 🏎️💨
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
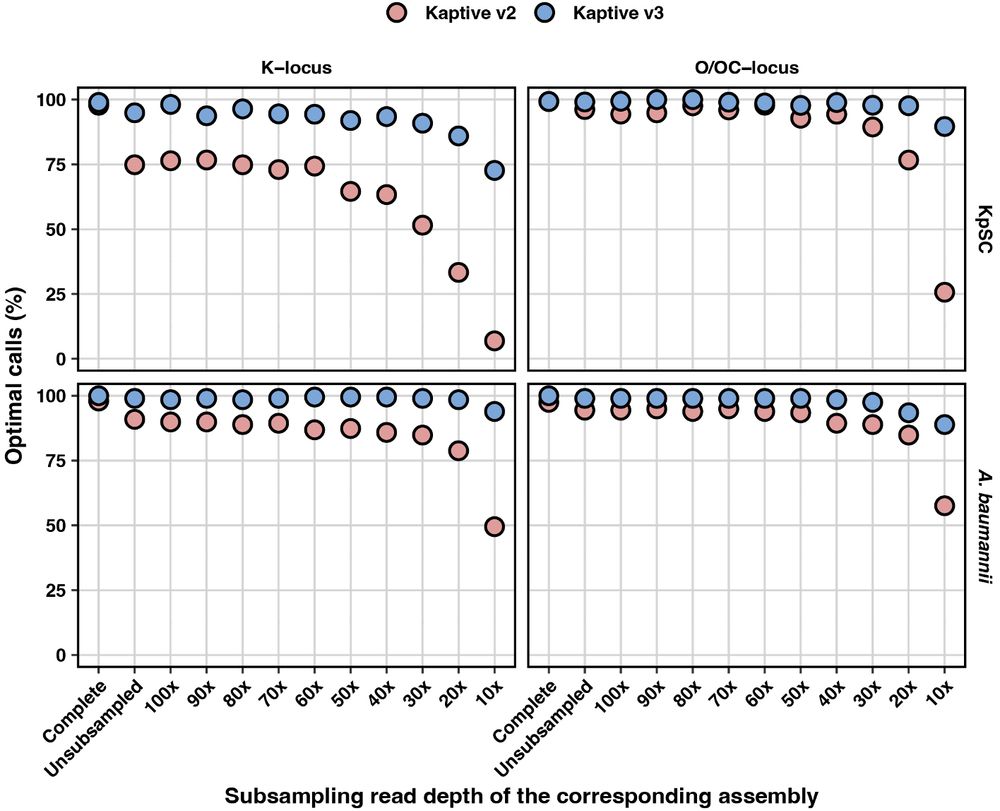
We then subsampled the corresponding short reads at decreasing depths and created sets of increasingly awful draft assemblies with loci broken over contigs and lots of genes missing.
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
We put together a special dataset specifically designed to test Kaptive. We had completed hybrid assemblies for KpSC (preprints.scielo.org/index.php/sc...) and A. baumannii (RefSeq).
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎

So enter Kaptive 3, a complete overhaul of Kaptive with a new algorithm designed to handle fragmented loci.
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
Because of how Kaptive 2 chose the best match locus, missing locus sequence resulted in a coverage bias for shorter loci in the database such, and could sometimes lead to inaccurate calls!
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
So in a nutshell, we traced Kaptive's issues with the Klebsiella K-locus all the way back to the gDNA, where:
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
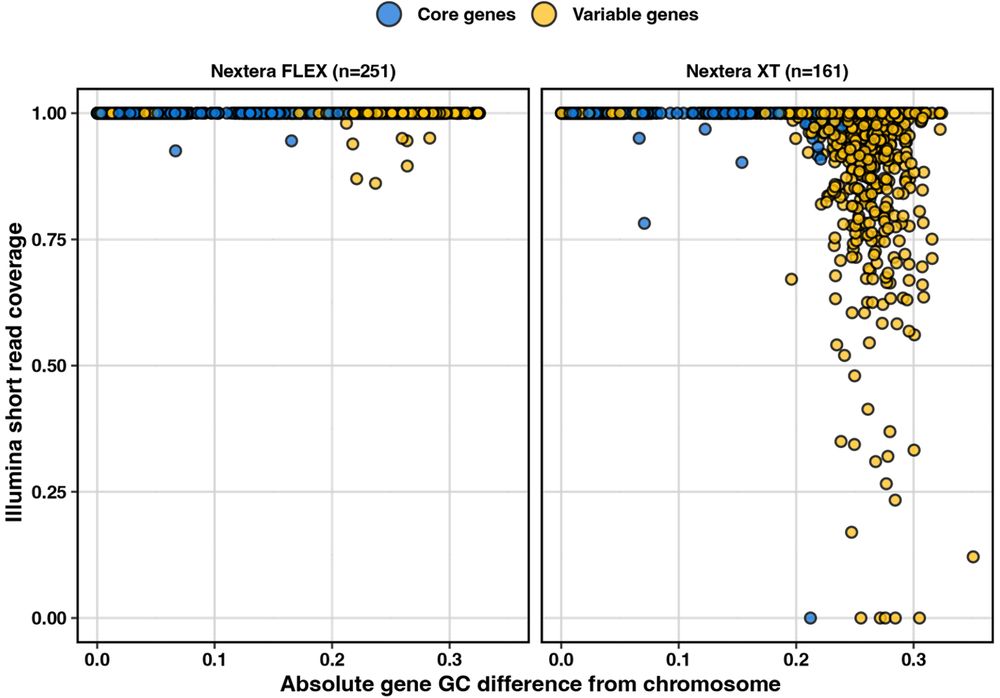
These genes have a very low GC compared to the rest of the Klebsiella chromosome, so we wondered if this was affecting how this part of the genome gets sequenced.
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
The major drivers of low confidence were K-loci that were 1) broken over contigs 🚫 and 2) missing genes ➡️➡️, events that were mostly co-occurring
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
So you may have noticed your Klebsiella K-locus results from previous versions of Kaptive (v1-2) having lots of untypeable calls ("Low" + "None" confidence) with draft assemblies; we certainly did!
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔

Super excited to finally present the preprint to accompany Kaptive 3 which we released last year!
Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...
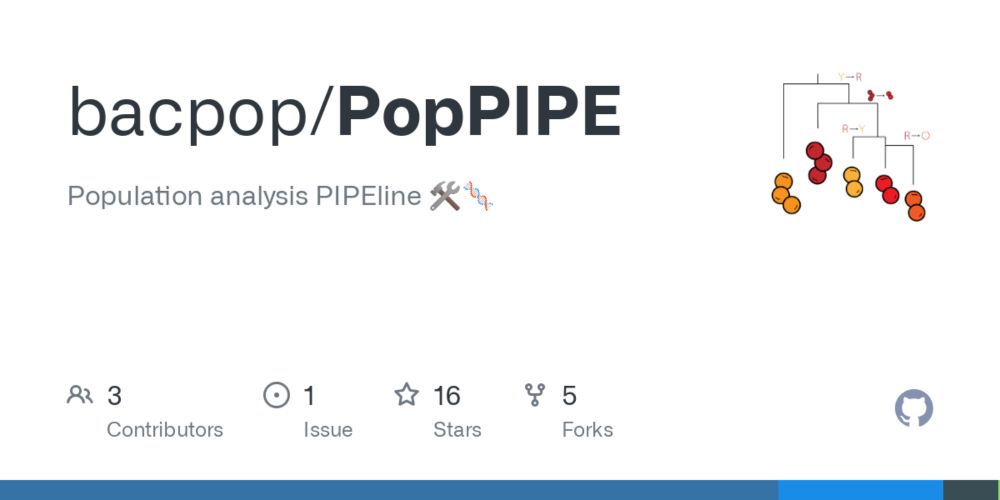
The PopPIPE (github.com/bacpop/PopPIPE) analysis pipeline can be used to subcluster data, create visualisations and run transmission analyses.
Preprint now here: www.biorxiv.org/content/10.1...
Including a case study on nosocomial transmission of vancomycin resistant Enterococcus faecium
whimsy driven development ✨
02.12.2024 01:53 — 👍 563 🔁 67 💬 22 📌 5Great talk by @bugsinyourguts.bsky.social: Raoultella is definitely Klebsiella + identified novel beta-lactamase variants in K. ornithinolytica/terrigena/planticola #Klebsiella2024
21.11.2024 09:22 — 👍 12 🔁 2 💬 1 📌 0Don't forget to join the #KlebClub Slack workspace!
#Klebsiella #KLEBS2024
join.slack.com/t/klebclub/s...
Lots of people are missing, let us know if you want to be included by replying!
You can also share to help us gain visibility
#MicroSky
go.bsky.app/EdereoU

A snapshot of my poster at Klebsiella 2024
If you are interested in how we make use of all of the data clinicians and scientists collect around the globe, come to poster 131 at this evening’s poster session! #klebs2024 #klebsiella2024
21.11.2024 12:56 — 👍 11 🔁 2 💬 0 📌 0
A small history lesson in #Klebsiella genomics in honour of #KLEBS24…
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/