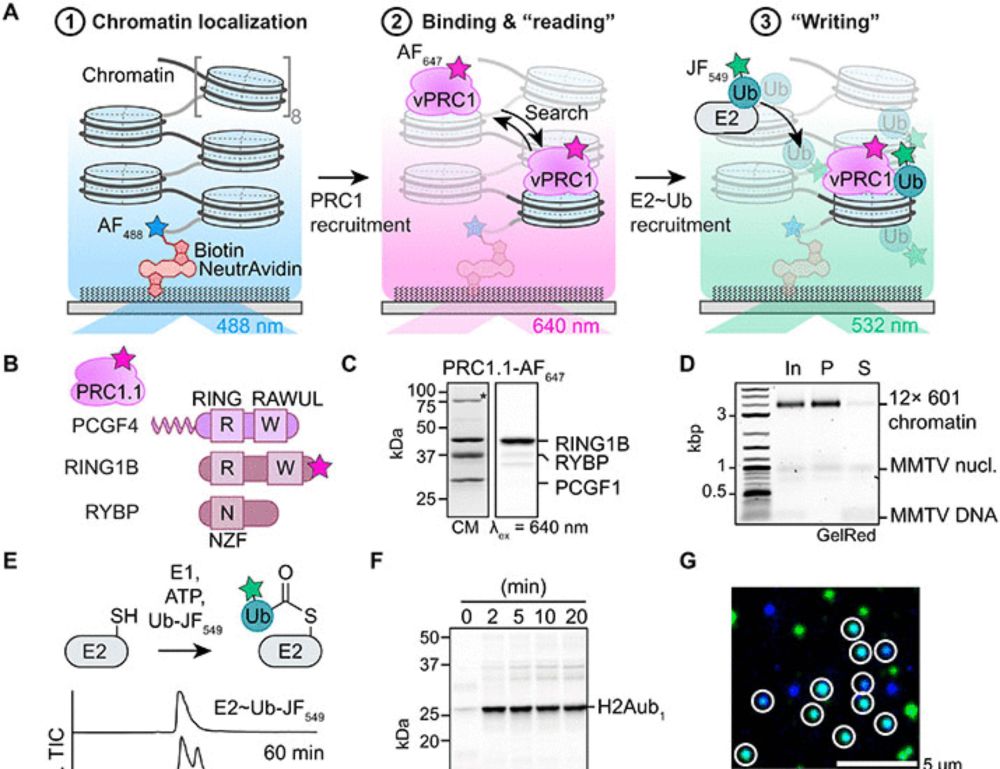
Single-molecule analysis reveals the mechanism of chromatin ubiquitylation by variant PRC1 complexes
Single-molecule experiments show that active conformation formation controls chromatin ubiquitylation kinetics by variant PRC1.
Our new study of chromatin ubiquitylation by variant PRC1 on the single-molecule scale: We visualize directly how vPRC1 ubiquitylates neighboring nucleosomes during a single binding event, showing a potential mechanism how H2Aub domains are established.
www.science.org/doi/10.1126/...
21.05.2025 18:59 —
👍 74
🔁 29
💬 2
📌 0
We are very grateful for the long-time financial support from the @erc.europa.eu, @snsf.ch, and EPFL (EDCH) who made this work possible! 🍀
22.05.2025 11:00 —
👍 1
🔁 0
💬 0
📌 0

Epigenetic landscape navigation by Polycomb complexes.
Using a single-molecule approach, we provide mechanistic insight into how Polycomb enzymes interpret chromatin context to control gene expression. In our study, we reveal that vPRC1 dynamically engages chromatin to form a catalytically competent complex with E2 and ubiquitin, a process governed by subtype-specific activity.
Using single-molecule dynamics, we observe real-time vPRC1 ubiquitylation of the chromatin fibers. vPRC1 dynamically engages chromatin, a process depending on E2 and H2Aub, while its PCGF subunit governs the formation of the catalytically competent complex with E2~Ub and enzymatic processivity.
22.05.2025 11:00 —
👍 2
🔁 0
💬 1
📌 0
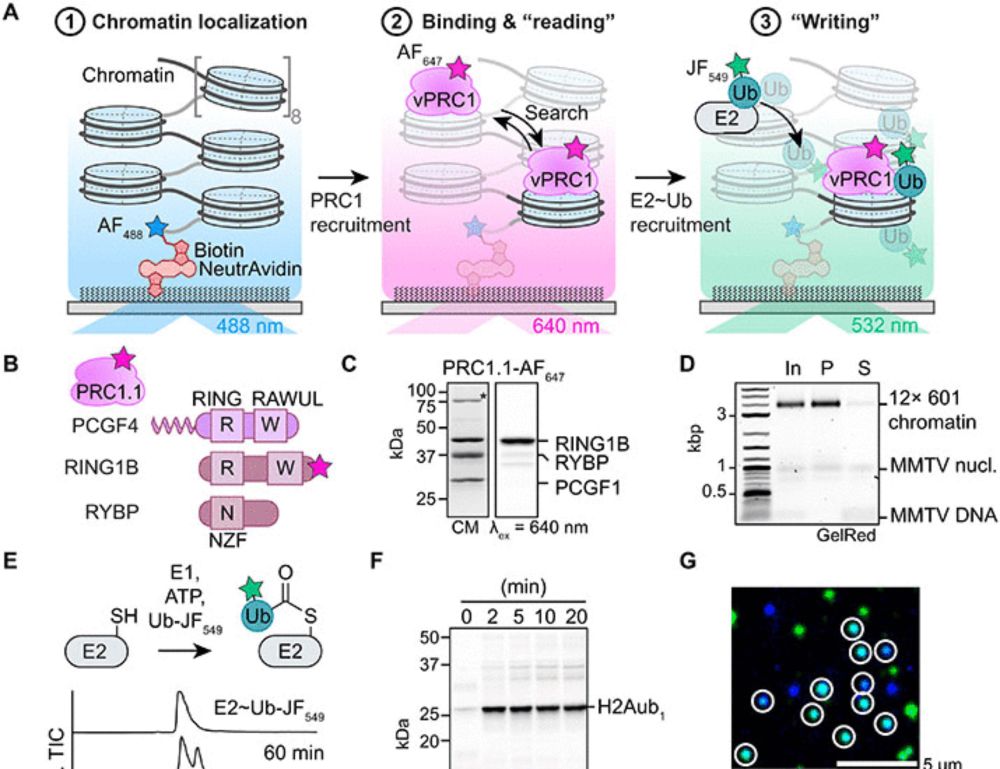
Single-molecule analysis reveals the mechanism of chromatin ubiquitylation by variant PRC1 complexes
Single-molecule experiments show that active conformation formation controls chromatin ubiquitylation kinetics by variant PRC1.
If you always wanted to know the kinetics of how the Polycomb enzymes modify chromatin, we are thrilled to share our work!!! 🎉📄🧬🔬
Thank you, @beatfierz.bsky.social for all the guidance as well as the @lcbm-epfl.bsky.social group members for all the support!
doi.org/10.1126/scia...
22.05.2025 11:00 —
👍 25
🔁 5
💬 1
📌 0
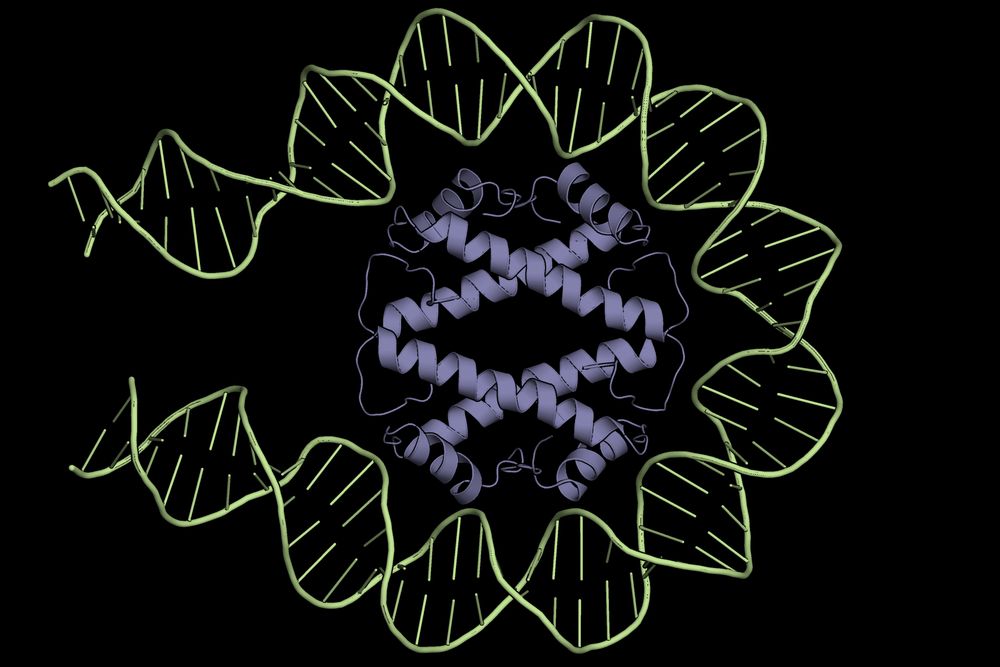
A snapshot of the HLp tetramer (purple) wrapping ~60 bp of DNA (green).
Excited to share our new preprint on HLp—a bacterial histone from Leptospira perolatii that forms stable tetramers and wraps ~60 bp of DNA: "DNA Wrapping by a Tetrameric Bacterial Histone" www.biorxiv.org/content/10.1...
@mpi-bio-fml.bsky.social
12.05.2025 09:16 —
👍 88
🔁 37
💬 4
📌 3
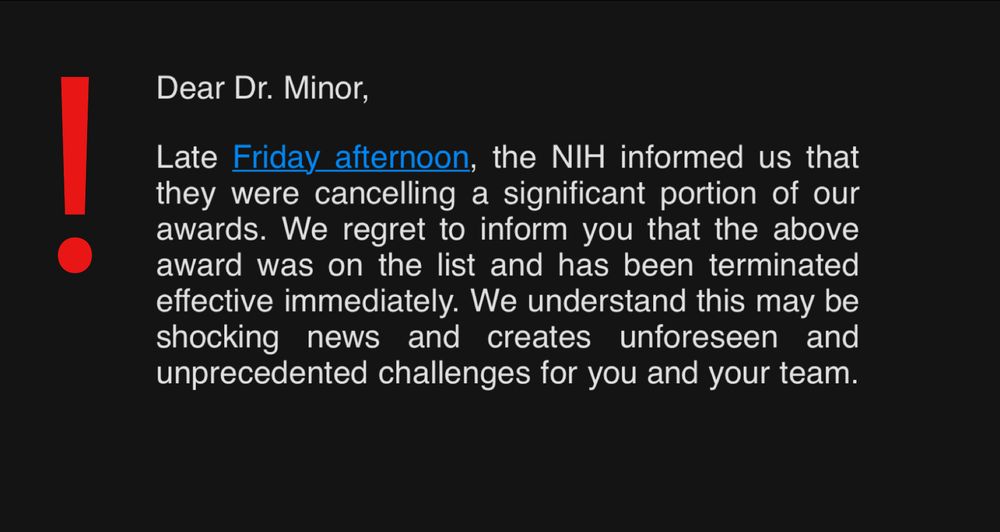
There are days in life that shake you.
I’m shattered 💔 to share that I just found out that the US Government terminated my 2024 NIH Director’s Early Independence Award (~$2 million), threatening my long-promised assistant professor job at Columbia University
& academic career... 1/🧵
18.03.2025 23:27 —
👍 1999
🔁 830
💬 93
📌 91
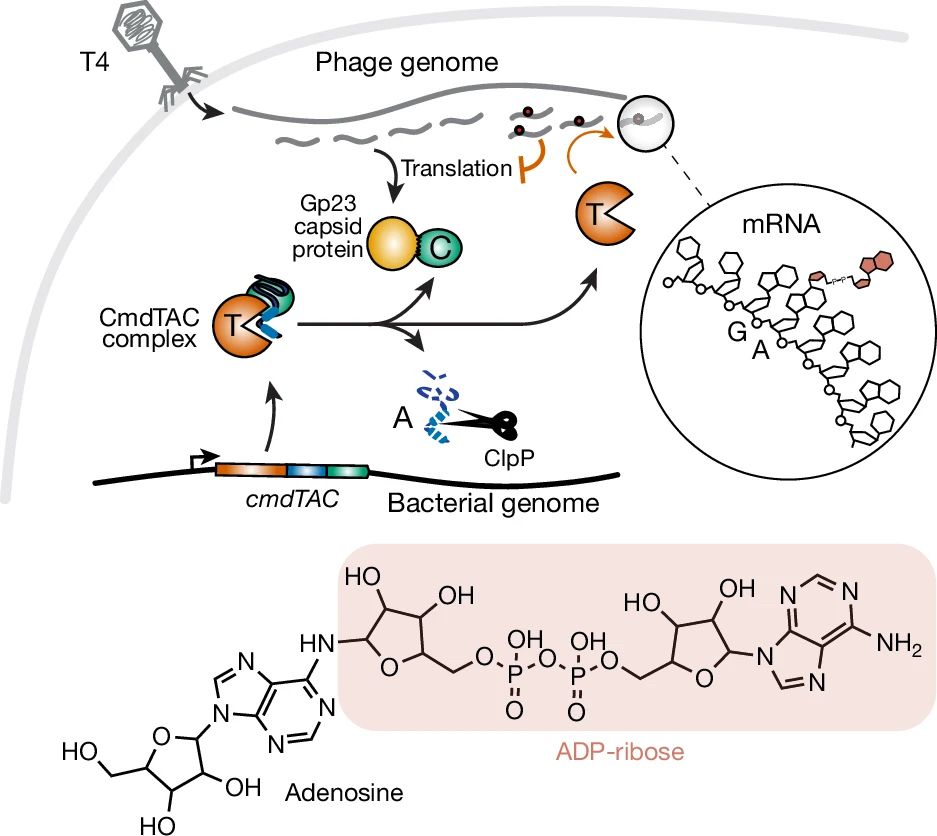
Graphic showing modification of mRNA with ADP-ribose mediated by bacterial enzyme cmdTAC
Check it out! Another novel RNA modification - ADP-ribosylation - that was previously only known on proteins. A cousin to #glycoRNA 👏
www.nature.com/articles/s41...
06.02.2025 19:38 —
👍 223
🔁 81
💬 3
📌 4
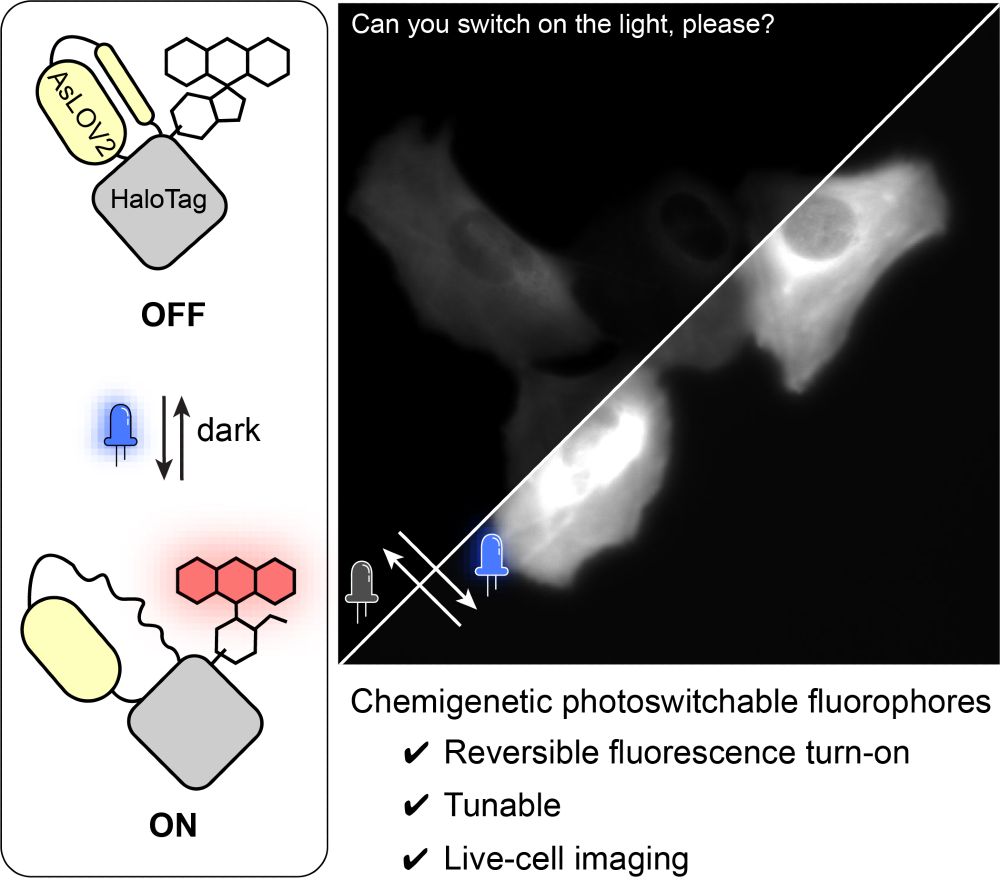
We made a photoswitchable HaloTag (psHaloTag), which can reversibly turn-on fluorogenic dyes upon illumination 💡. Congrats to Franzi, Bego and all co-authors, check out our preprint below 👇
www.biorxiv.org/content/10.1...
07.01.2025 08:45 —
👍 206
🔁 52
💬 2
📌 2
A screenshot of a paper on bioarxiv illustrating the lack of blue sky share button!
Would you like to see @biorxivpreprint.bsky.social add a share to blue sky button?! I know I would! Share this post to let @richardsever.bsky.social @erictopol.bsky.social and others at bioarxiv know!
02.12.2024 12:10 —
👍 384
🔁 196
💬 9
📌 6


🥳 Today we celebrated the PhD defense of our very first PhD student Kevin Schiefelbein 🤩 Kevin told us about his Lasso Peptide journey over the past four years, and it was so great to see the full story coming together. Congratulations Kevin, and thank you for everything! #proudPI
04.12.2024 20:45 —
👍 26
🔁 3
💬 0
📌 0
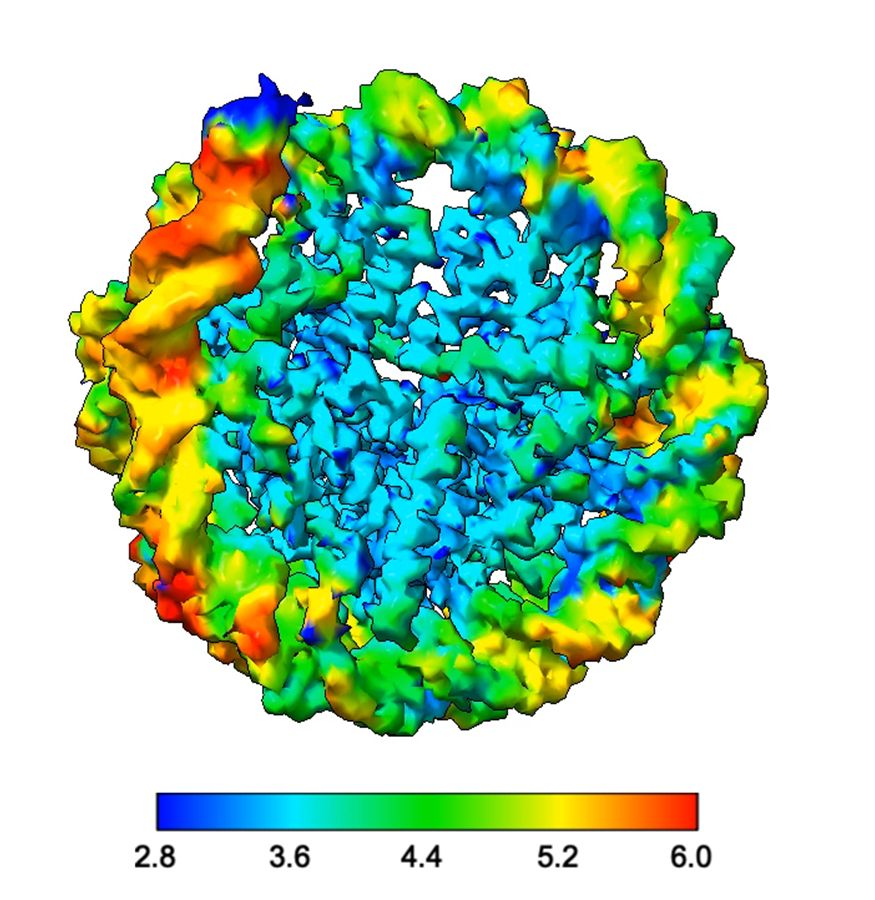
behold our paper on Medusavirus nucleosomes. Fantastic work by Chelsea Toner, @nhoitsma.bsky.social, and Sashi Weerawarana This giant virus also encodes a linker histone, more about that in the paper 🧪👩🔬 🧵.
www.nature.com/articles/s41...
23.10.2024 18:19 —
👍 149
🔁 53
💬 5
📌 4
First attempt in a (loosely defined) Single-Molecule Biophysics starter pack. Please share and (self-) nominate. go.bsky.app/CteFZM5
13.11.2024 08:23 —
👍 107
🔁 60
💬 65
📌 2
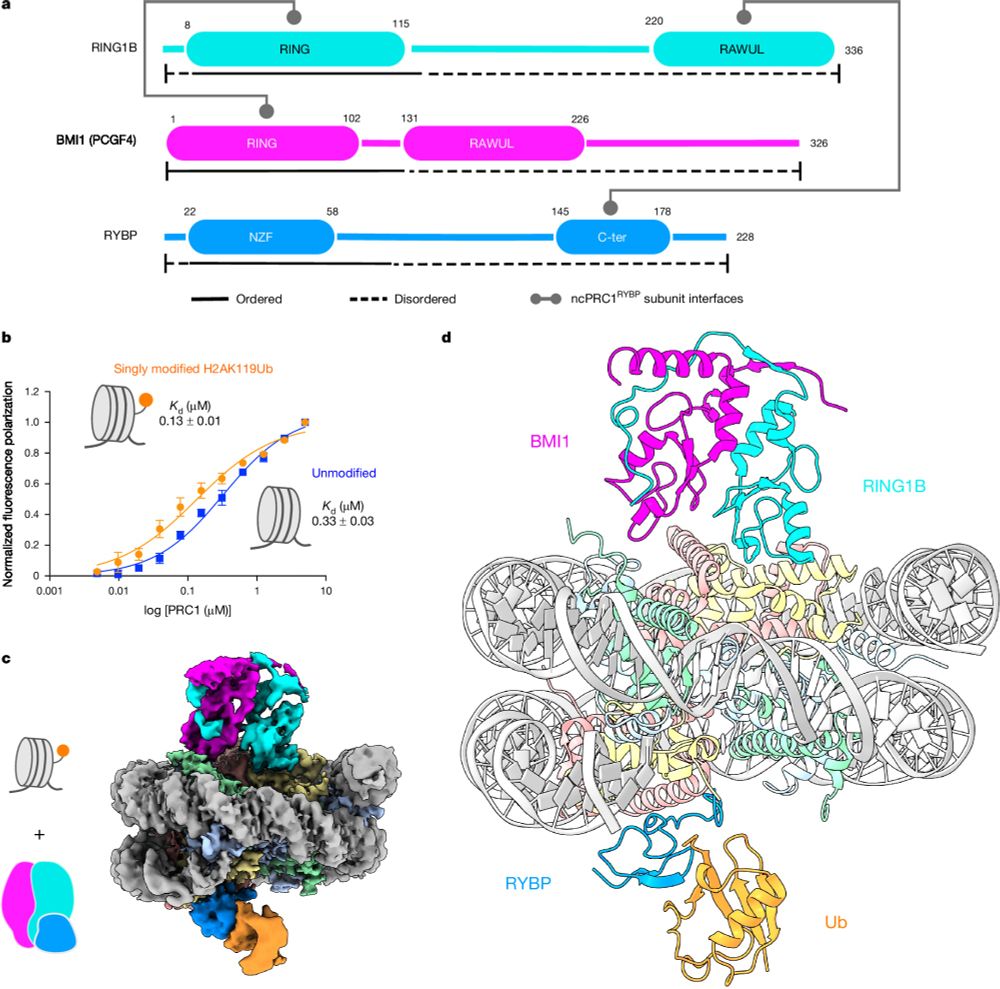
Read–write mechanisms of H2A ubiquitination by Polycomb repressive complex 1
Nature - Cryo-electron microscopy and biochemical studies elucidate the read–write mechanisms of non-canonical PRC1-containing RYBP in histone H2A lysine 119 monoubiquitination and their...
Please see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator JP Armache! Also big thanks to Mark Foundation for Cancer Research for the support! rdcu.be/dZ5HZ
13.11.2024 18:23 —
👍 115
🔁 31
💬 7
📌 4
📯📯 Chemical Biology starter pack 📯📯
140 PIs & groups posting primary research in chemical biology.
Please reskeet (?) & reply to this post if you want to be added!
go.bsky.app/KLrTWoj
Let's make ChemBioSky even greater again!
#realtimechem #FluorescenceFriday #chemsky #chemtwitter #AgentOrange
17.11.2024 22:58 —
👍 139
🔁 115
💬 48
📌 8
Check out this list of ubiquitin and Ubl profiles to follow. Repost if you want me to add you to it. Let’s create a good community here ☁️ go.bsky.app/3VPQofn
15.11.2024 19:38 —
👍 49
🔁 48
💬 6
📌 1

⚠️THREAD OF ALL SCIENCE-RELATED STARTER PACKS! ⚠️
🔄Share to help people finding its topic/field
- STRUCTURAL BIOLOGY
Structural Biology 1: bsky.app/starter-pack...
Structural Biology 2: bsky.app/starter-pack...
Crystallography: bsky.app/starter-pack...
Synchrotron&CryoEM: bsky.app/starter-pack...
17.11.2024 18:48 —
👍 287
🔁 180
💬 39
📌 20
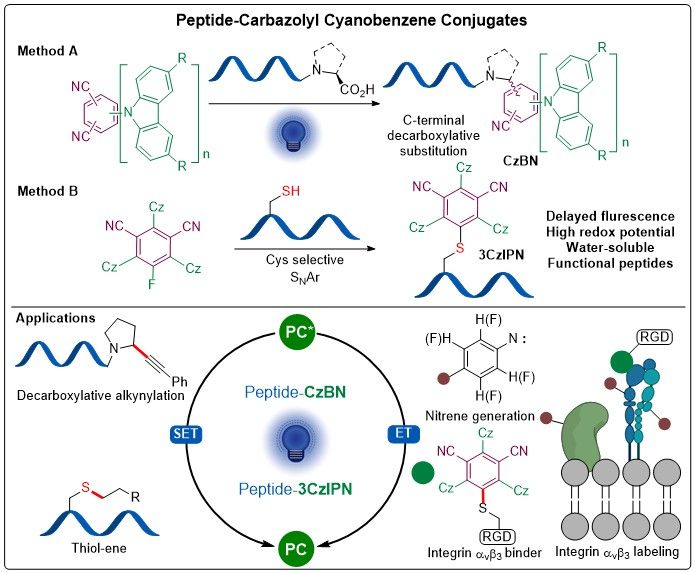
On November 20, the new class of peptide-cyanobenzene dye photocatalysts developed by Xingyu have been first made public in a preprint at ChemRxiv, including a great collaboration Wei Cai in the @beatfierz.bsky.social group and Dr. Anne Sophie Chauvin
chemrxiv.org/engage/chemr...
23.11.2024 08:29 —
👍 5
🔁 2
💬 0
📌 0