Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling - preLights
Ready, set, brominate: RebH and engineered variant for bromination of peptidyl-Trp residues
Some excellent preLights to end 2025 🎆
The latest was written by Zhang-He Goh @goh-zhanghe.bsky.social, discussing an enzymatic method for tryptophan-specific bromination described in a recent #preprint from the lab of @amyweeks.bsky.social.
#preLight ⬇️
prelights.biologists.com/highlights/e...
31.12.2025 14:14 — 👍 3 🔁 1 💬 0 📌 1

Great work by recently minted PhD Haley Bridge and recent BS Chase Radziej! We are grateful to the NIH for supporting this work. Public investment in fundamental science fuels innovation and is worth defending.
21.12.2025 03:58 — 👍 3 🔁 0 💬 0 📌 0

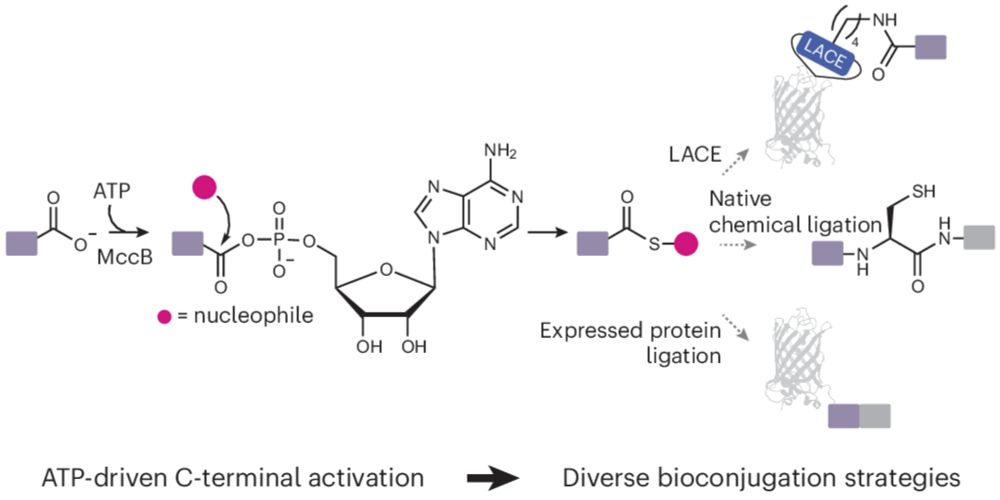
We found that 4V produced higher conversion and had expanded sequence scope compared to RebH. We took advantage of this newly revealed activity to combine enzymatic bromination with Suzuki-Miyaura cross-coupling to enable late-stage chemoenzymatic functionalization of unmodified bioactive peptides.
21.12.2025 03:58 — 👍 0 🔁 0 💬 1 📌 0
The observation of peptide activity in RebH motivated us to examine the RebH variant 4V, which was engineered by @jclewislab.bsky.social to accommodate larger substrates 👇https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201411901
21.12.2025 03:58 — 👍 1 🔁 0 💬 1 📌 0

New preprint! We found that the flavin-dependent halogenase RebH catalyzes sequence-tolerant Trp bromination in peptides 🧪https://www.biorxiv.org/content/10.64898/2025.12.17.694899v1
21.12.2025 03:58 — 👍 33 🔁 14 💬 2 📌 0
New in ACS SynBio: led by Dennis Bolshakov, we used the awesome power of yeast to define how expression levels, noise, and sequence program the dynamics of synthetic protein waves, allowing us to genetically encode new cellular timescales stable over generations!
pubs.acs.org/doi/full/10....
17.12.2025 15:24 — 👍 27 🔁 11 💬 2 📌 1
Thrilled to share our work on the 🔥 single-celled predator Podophrya collini, which rewires its cell morphology to hunt more efficiently. Huge thanks to our amazing team—Amy, Lauren, Omaya, Marine, Mari, and especially Scott—for making this shine! ✨
18.11.2025 17:12 — 👍 29 🔁 13 💬 1 📌 0
How do cells adapt morphology to function? In a 🔥 preprint by @zjmaggiexu.bsky.social , with @dudinlab.bsky.social and @amyweeks.bsky.social , we identify a self-organizing single-cell morphology circuit that optimizes the feeding trap structure of the suctorian P. collini. 🧵 tinyurl.com/4k8nv926
18.11.2025 16:15 — 👍 131 🔁 55 💬 4 📌 11
A beautiful example of how spatiotemporal dynamics can enable multiplexed measurements.
16.09.2025 11:36 — 👍 6 🔁 2 💬 1 📌 0
DNA-Scaffolded Ultrahigh-Throughput Reaction Screening
Discovering and optimizing reactions is central to synthetic chemistry. However, chemical reactions are traditionally screened using relatively low-throughput methods, prohibiting exploration of diver...
Excited to share our new preprint, which was years in the making! chemrxiv.org/engage/chemr...
New reactions are typically developed by trial and error. How can we speed up this process? Read on to learn how we used DNA scaffolding to perform >500,000 parallel reactions on attomole scale.
1/n
14.08.2025 17:40 — 👍 40 🔁 19 💬 1 📌 3

A handmade poster that says "We are all in this together" with a DNA double helix in the center.
Congratulations to first author Kasia Radziwon and co-authors Laura Campbell, Lauren Mazurkiewicz, Sopo Jalalshvili, Izabelle Eppinger, and Aanika Parikh! We are grateful to the NIH for supporting this work. Public investment in fundamental science fuels innovation and is worth defending.
13.08.2025 17:20 — 👍 7 🔁 0 💬 0 📌 0

A crystal structure of the OspF homolog SpvC bound to a substrate peptide. Plots of % sites beta-eliminated for pThr and pSer with several OspF variants.
Leveraging the throughput of our approach, we took a deep dive into the molecular basis of OspF specificity. We identified enzyme residues in the L12 loop that influence OspF’s specificity for pThr vs. pSer and were able to shift this specificity through protein engineering
13.08.2025 17:20 — 👍 3 🔁 0 💬 1 📌 0

OspF is a pThr lyase from Shigella flexneri that targets MAPKs in innate immune signaling. We profiled OspF activity on PhosPropels and found that its catalytic domain has an intrinsic preference for the the MAPK activation loop motif pThr-X-pTyr.
We also found that the catalytic domain of the pThr lyase OspF (a functionally and mechanistically distinct phosphoeraser from Shigella flexneri) has and intrinsic preference for the pThr-Xaa-pTyr motif found in Erk and p38 MAP kinase activation loops that does not depend on its MAPK docking motif
13.08.2025 17:20 — 👍 0 🔁 0 💬 1 📌 0

The Legionella pneumophila phosphatase WipB targets the lysosomal nutrient sensing machinery. WipB disfavors substrates with Pro at +1 and phosphosites flanked by acidic residues.
We applied our approach to define sequence motifs for 8 phosphoerasers spanning diverse species, folds, and enzymatic mechanisms. Example: The Legionella pneumophila phosphatase WipB uses multiple selectivity filters, disfavoring substrates with Pro at +1 and phosphosites flanked by acidic residues
13.08.2025 17:20 — 👍 1 🔁 0 💬 1 📌 0

Workflow for PhosPropel-based profiling of phosphoerasers. Cells are lysed, protein is digested, and phosphopeptides are enriched to generated a phosphoproteome-derived peptide library (PhosPropel). The PhosPropel can then be treated with a phosphoeraser enzyme and analyzed using LC-MS/MS to profile enzyme specificity.
We developed an LC-MS/MS-based in vitro assay for dephosphorylation of human phosphoproteome-derived peptide libraries (PhosPropels). We use statistical comparison of phosphopeptide sequence features in enzyme-treated samples vs. controls for deep specificity profiling of phosphoeraser enzymes.
13.08.2025 17:20 — 👍 0 🔁 0 💬 1 📌 0

Position-specific frequency matrices can be used to calculate z-scores comparing enzyme-treated and control samples. The z-scores are plotted as heatmaps that represent an enzyme specificity profile.
New preprint: we developed a method that uses phosphoproteome-derived peptide libraries (PhosPropels) for deep specificity profiling of phosphatases and phospholyases www.biorxiv.org/content/10.1...
13.08.2025 17:20 — 👍 48 🔁 19 💬 1 📌 2

OspF is a pThr lyase from Shigella flexneri that dampens innate immune signaling. We treated PhosPropels with OspF and found that this enzyme has intrinsic specificity for the pThr-X-pTyr motif of MAPK activation loops independent of an N-terminal MAPK docking motif.
We also found that the catalytic domain of the pThr lyase OspF (a functionally and mechanistically distinct phosphoeraser from Shigella flexneri) has and intrinsic preference for the pThr-Xaa-pTyr motif found in Erk and p38 MAP kinase activation loops that does not depend on its MAPK docking motif
13.08.2025 17:02 — 👍 0 🔁 0 💬 0 📌 0

WipB is a Ser/Thr phosphatase from Legionella pneumophila that targets lysosomal nutrient sensing machinery. PhosPropel analysis revealed that WipB disfavors substrates with Pro at +1 and phosphosites flanked by acidic residues.
We applied our approach to define sequence motifs for 8 phosphoerasers spanning diverse species, folds, and enzymatic mechanisms. Example: The Legionella pneumophila phosphatase WipB uses multiple selectivity filters, disfavoring substrates with Pro at +1 and phosphosites flanked by acidic residues
13.08.2025 17:02 — 👍 1 🔁 0 💬 1 📌 0

Workflow for phosphoeraser specificity profiling. Cells are lysed, protein is digested, phosphopeptides are enriched, the phosphopeptide library is treated with an eraser enzyme, and phosphosites are analyzed by LC-MS/MS
We developed an LC-MS/MS-based assay for dephosphorylation of human phosphoproteome-derived peptide libraries (PhosPropels). We use statistical comparison of phosphopeptide sequence features in enzyme-treated samples vs. controls for deep specificity profiling of phosphoerasers.
13.08.2025 17:02 — 👍 0 🔁 0 💬 1 📌 0

SynchroSep-MS: Parallel LC Separations for Multiplexed Proteomics
Achieving high throughput remains a challenge in MS-based proteomics for large-scale applications. We introduce SynchroSep-MS, a novel method for parallelized, label-free proteome analysis that leverages the rapid acquisition speed of modern mass spectrometers. This approach employs multiple liquid chromatography columns, each with an independent sample, simultaneously introduced into a single mass spectrometer inlet. A precisely controlled retention time offset between sample injections creates distinct elution profiles, facilitating unambiguous analyte assignment. We modified the DIA-NN workflow to effectively process these unique parallelized data, accounting for retention time offsets. Using a dual-column setup with mouse brain peptides, SynchroSep-MS detected approximately 16,700 unique protein groups, nearly doubling the peptide information obtained from a conventional single proteome analysis. The method demonstrated excellent precision and reproducibility (median protein %RSDs less than 4%) and high quantitative linearity (median R2 greater than 0.96) with minimal matrix interference. SynchroSep-MS represents a new paradigm for data collection and the first example of label-free multiplexed proteome analysis via parallel LC separations, offering a direct strategy to accelerate throughput for demanding applications such as large-scale clinical cohorts and single-cell analyses without compromising peak capacity or causing ionization suppression.
Check out our new manuscript on parallel LC separations! Super cool how the very high scan rates of modern MS systems coupled with DIA can allow us to run several samples at the same time with little loss in depth. Congrats to Noah and the team. #JASMS pubs.acs.org/doi/10.1021/...
30.07.2025 17:36 — 👍 6 🔁 3 💬 1 📌 0
Bravo to Clara Frazier, Debashrito Deb
@thepeptidetailor.bsky.social and coauthors Will Leiter @wleiter1999.bsky.social and Umasankar Mondal!
18.07.2025 14:42 — 👍 1 🔁 0 💬 0 📌 0

Great success at the Ono Pharma Foundation Symposium in Boston! Highlights include inspiring talks by Xiao Wang @amyweeks.bsky.social Robert Spitale @stevenbanik.bsky.social @michael-erb.bsky.social Matthew Shoulders Michelle Arkin and a keynote from Chuan He. Posters fueled inspired exchange.
02.07.2025 01:31 — 👍 3 🔁 1 💬 1 📌 0

Debashrito Deb holding up his poster award at the 2025 Bioorganic GRC
Congrats to Weeks lab graduate student Debashrito Deb @thepeptidetailor.bsky.social, who won a poster prize at the Bioorganic GRC last week! Thanks to conference chairs @doc-jlmeier.bsky.social and Denise Field who knocked it out of the park with a memorable and inspiring meeting this year!
26.06.2025 15:44 — 👍 27 🔁 4 💬 2 📌 1
Despite ~20 years in/around #chembio research, I went to my first Bioorganic GRC this week. This community is amazing and so supportive. I feel energized (and tired, lol) and find myself rooting for the next generation of chemical biologists. Sooooo much awesome science - We can’t/won’t be stopped!
21.06.2025 16:43 — 👍 33 🔁 5 💬 0 📌 2

The 13th General Meeting of the International Proteolysis Society will be held in Búsios, Brazil Oct 26-30, 2025. Training workshops will be held at the Instituto Oswaldo Cruz Oct 23-23. Register now! Links below.
1/3
22.05.2025 04:22 — 👍 7 🔁 8 💬 1 📌 1

excited to share our latest work now online
@natmetabolism.nature.com, led by
@kyle-flickinger.bsky.social, where we unravel a mechanistic basis for the conditional essentiality of NADK, one of the many interesting hits from our previously reported CRISPR screening with HPLM rdcu.be/ekpu6
02.05.2025 15:32 — 👍 59 🔁 19 💬 17 📌 0
MIT biology lab using computational and experimental methods to study protein structure, function, and interactions
Professor of Political Science & Public Affairs & Director of European Studies, University of Wisconsin - Madison. International political economy, international relations, & international finance. 🇺🇸 & 🇪🇺 politics & economic policy. Tradeoffs in everything.
Scientist exploring how AI can be used to accelerate discovery and innovation in biology. Creator of SciBud Media
Associate Professor at UCSF, visiting scientist at the Gladstone Institutes. Expertise in mass spectrometry and proteomics. Views are my own. profiles.ucsf.edu/danielle.swaney
Chemical Biologist & Research Groupleader at Max Planck & TU Dortmund. Interested in DUBs, proteases, ubiquitin, the outdoors and more. Views are my own. cgc.mpg.de/gersch
Molecular biologist using functional genomics. Started with C. elegans, then diversified.
graduate student - University of Wisconsin Madison
scientist at UC Berkeley inventing advanced genomic technologies
lover of molecules, user of computers
https://scholar.google.com/citations?user=63ZRebIAAAAJ&hl=en
Chemical Biologist. Tenure track assistant prof @EPFL. Trying to make sense of the world by changing one variable at a time. https://www.epfl.ch/labs/libn/
Student-run account for the Lingjun Li Lab at UW-Madison. #TeamMassSpec @uwmadpharmacy @uwmadisonchem
Omics and data science. Track new research with AI using rescoop.xyz /
Assistant Professor at Cedars-Sinai. Opinions are my own and do not reflect my employer
Mass spectrometry for quantitative proteomics. The Olsen group at the Center for Protein Research, University of Copenhagen.
Science / Mass Spectrometry
Bringing Proteoforms to Life (sm)
Our Mission is to promote collaboration, education, and innovative research that will accelerate the comprehensive analysis of all human proteoforms. ctdp.org
Welcome to the Sieber group BlueSky page. Here we keep you up to date on our research (and other interesting things) at the TUM.⚗️🧪🧫🧬🔬👩💻
Our homepage: https://www.bio.nat.tum.de/oc2/home/
Our LinkedIn: https://www.linkedin.com/company/sieber-lab
PhD candidate, UW-Madison
Li Research Group & Ricke Lab
Computational biologist; Associate Prof. at University of Wisconsin-Madison; Jeanne M. Rowe Chair at Morgridge Institute
British, But In Las Vegas and NYC
ezitron.76 Sig
Newsletter - wheresyoured.at
https://linktr.ee/betteroffline - podcast w/ iheartradio
Chosen by god, perfected by science
CEO at EZPR.com - Award-Winning Tech PR