
We are happy to present a new paper on gastric tumour evolution in response to peri-operative chemotherapy. www.biorxiv.org/content/10.1... 🧵 1/6
31.07.2025 06:27 — 👍 13 🔁 6 💬 1 📌 0@kezc.bsky.social
Lecturer in cancer bioinformatics at Cardiff University. https://github.com/kcleal

We are happy to present a new paper on gastric tumour evolution in response to peri-operative chemotherapy. www.biorxiv.org/content/10.1... 🧵 1/6
31.07.2025 06:27 — 👍 13 🔁 6 💬 1 📌 0Nice work! Do you think it would be hard to calculate perplexity for short read data?
03.07.2025 07:05 — 👍 0 🔁 0 💬 1 📌 0
New work from the lab trying to wrap our heads around the massive complexity of the human transcriptome revealed by long-read RNA-seq! Fun collab with Gloria Sheynkman. www.biorxiv.org/content/10.1...
02.07.2025 23:46 — 👍 55 🔁 22 💬 2 📌 0Thank you, Francisco 🙏
26.06.2025 21:08 — 👍 1 🔁 0 💬 0 📌 0Thanks Wales Bioinformatics Network 🙂
26.06.2025 16:13 — 👍 1 🔁 0 💬 0 📌 0
🚨 Our GW paper is out in Nature Methods!🥲
GW is a fast genomics browser (up to 100x faster!)
github.com/kcleal/gw
Also, just released a Python interface for GW
github.com/kcleal/gwplot
📝 nature.com/articles/s4159…
#Genomics #Bioinformatics
Very impressive work 👏
10.04.2025 16:22 — 👍 1 🔁 0 💬 0 📌 0A really useful resource
08.04.2025 07:49 — 👍 3 🔁 2 💬 0 📌 0@adbeggs.bsky.social Hi Andrew. Fantastic talk today at the WCRC conference! It was very inspiring 👏
03.03.2025 19:13 — 👍 1 🔁 0 💬 1 📌 0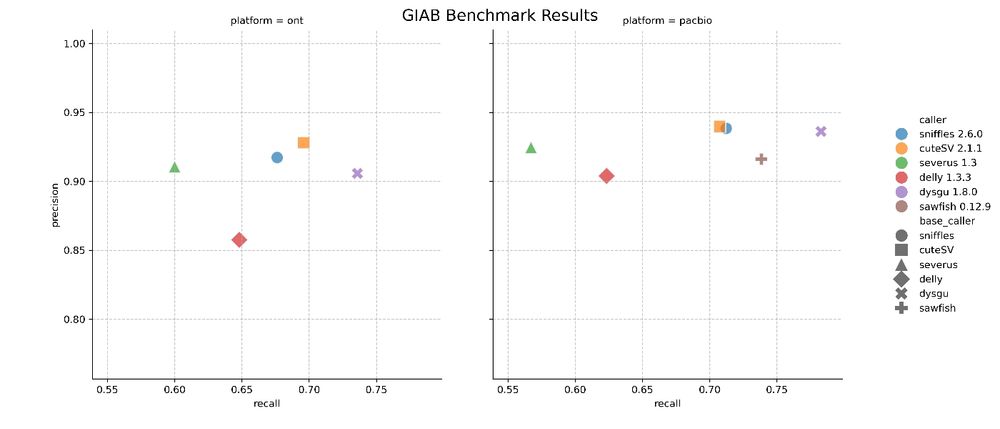
🧬🖥️ Just released Dysgu v1.8 which now supports phasing of structural variants when using long reads. Makes SV calling much more sensitive - see PacBio benchmark 👉 github.com/kcleal/SV_Be... dysgu repo github.com/kcleal/dysgu...
26.02.2025 11:37 — 👍 8 🔁 4 💬 0 📌 0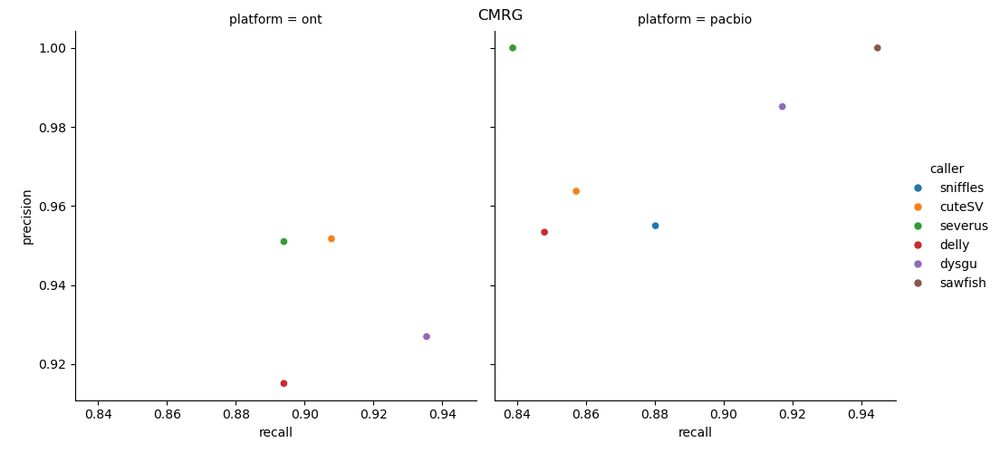 29.11.2024 14:49 — 👍 0 🔁 0 💬 0 📌 0
29.11.2024 14:49 — 👍 0 🔁 0 💬 0 📌 0
 29.11.2024 14:48 — 👍 0 🔁 0 💬 0 📌 0
29.11.2024 14:48 — 👍 0 🔁 0 💬 0 📌 0
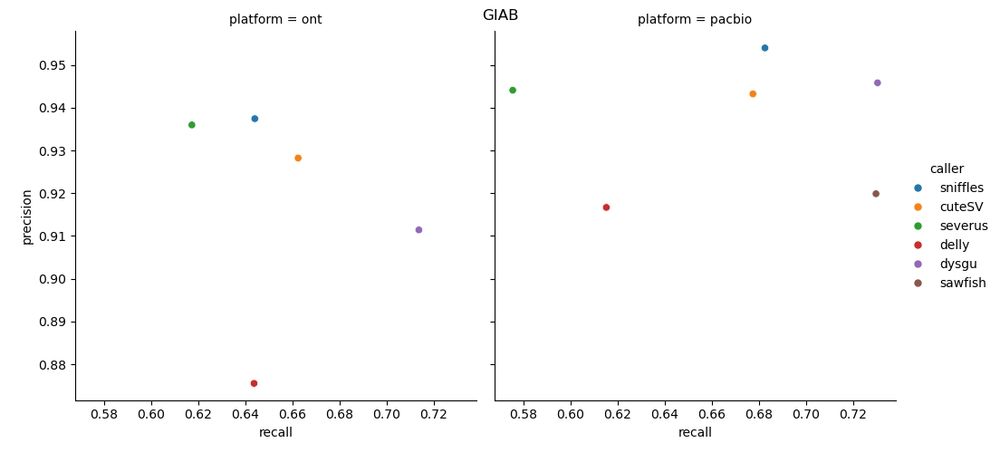
🧬🖥️ Sharing an update to my SV benchmark repo. More callers have been added for the CMRG/GIAB benchmarks. github.com/kcleal/SV_Be.... Also now uses #Nextflow and tests the newest #PacBio Vega data, along with #ONT kit14. Results are on the GitHub page 👉
29.11.2024 13:40 — 👍 11 🔁 2 💬 2 📌 0🧬 🖥️
18.11.2024 20:22 — 👍 0 🔁 0 💬 0 📌 0Also my first foray into rust land - lots to like I have to say!
18.11.2024 16:39 — 👍 0 🔁 0 💬 0 📌 0
Hi all. Just released a fast library (C++/Rust/Python) for performing interval intersections. Uses a novel superset-index that tends to be faster than current libs, and also has a SIMD accelerated counting algorithm 👇 github.com/kcleal/super...
18.11.2024 15:44 — 👍 18 🔁 5 💬 4 📌 0Thanks 👍 Could I be added? Cheers
12.11.2024 21:57 — 👍 0 🔁 0 💬 1 📌 0Whole genome in 2s 😂 wow👏
12.11.2024 21:20 — 👍 1 🔁 0 💬 1 📌 0Hi @robp.bsky.social, not sure if I qualify, but could I be added to the list? Cheers 👍
12.11.2024 16:02 — 👍 1 🔁 0 💬 1 📌 0I made a starter pack for algorithmic genomics. It's certainly incomplete, but already has a ton of awesome peeps. Let me know if you know people I should add (with a focus on algorithms and data structures in genomics)
go.bsky.app/TRWCnZs

BioNumpy looks really good, looking forward to giving this a try www.nature.com/articles/s41...
11.11.2024 19:05 — 👍 1 🔁 1 💬 0 📌 0
I created a genomics+bioinformatics starter pack. If I left you off, *please* reply and I'll add you! go.bsky.app/B5YYBfq
22.10.2024 12:34 — 👍 176 🔁 106 💬 99 📌 7Hello world! This looks promising
13.09.2023 22:44 — 👍 4 🔁 0 💬 0 📌 0