PhD Student – Structural Biology of Actin Filament Assemblies | Werken bij AVL
Are you excited to uncover the molecular mechanisms of actin filament assemblies that drive cell migration using cryo-EM?
Only 1.5 week to apply for this exciting PhD position in my newly established lab at the Netherlands Cancer Institute in Amsterdam: www.werkenbijavl.nl/vacatures/ph...
28.10.2025 09:17 — 👍 6 🔁 7 💬 0 📌 0

In our new article, we show how P. aeruginosa, a major cause of chronic respiratory infections in #cysticfibrosis (CF), uses the Type VI Secretion System #T6SS and specific #toxins to eliminate competing bacteria
www.cell.com/cell-reports...
#cryoEM @dshatskiy.bsky.social @jakecolautti.bsky.social
14.09.2025 08:52 — 👍 33 🔁 14 💬 1 📌 0
Thank you!
04.09.2025 19:45 — 👍 1 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 19:45 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 19:45 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 18:25 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 13:48 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 13:38 — 👍 0 🔁 0 💬 0 📌 0
Thank you!
04.09.2025 12:58 — 👍 0 🔁 0 💬 0 📌 0
Thrilled to be one of 12 awardees of the prestigious #ERCStG from the LS1 panel this year! Over the next five years, we’ll study how intracellular bacterial pathogens #Listeria and #Shigella manipulate #actin to spread infection, with the aim of finding ways to stop them. Exciting times ahead!
04.09.2025 12:38 — 👍 31 🔁 4 💬 7 📌 0
We bought a nail dryer from Amazon for 20 euro 🙂
14.08.2025 08:20 — 👍 1 🔁 0 💬 0 📌 0
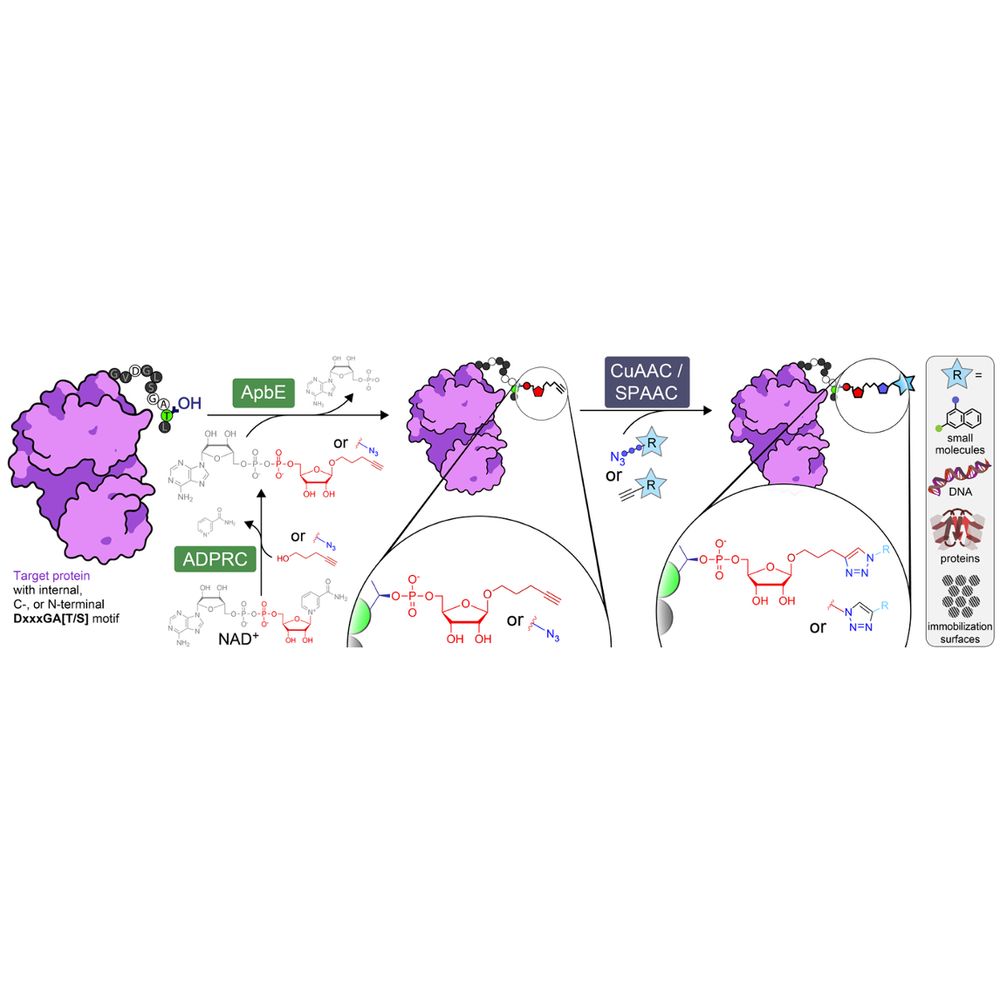
Overview of ADD-tagging including ADP-ribosyl cyclase (ADPRC)-catalysed dinucleotide substrate generation and two step chemoenzymatic labelling of target proteins with the flavin transferase ApbE, followed by click chemistry-based functional group attachment.
🚨 preprint 2️⃣ this month: our (purely experimental🧪) venture into #ChemBio
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
20.05.2025 12:29 — 👍 54 🔁 15 💬 4 📌 2
Great work by @jakecolautti.bsky.social @dshatskiy.bsky.social Eileen Bates, Nathan P. Bullen & John C. Whitney
01.05.2025 15:37 — 👍 0 🔁 0 💬 0 📌 0
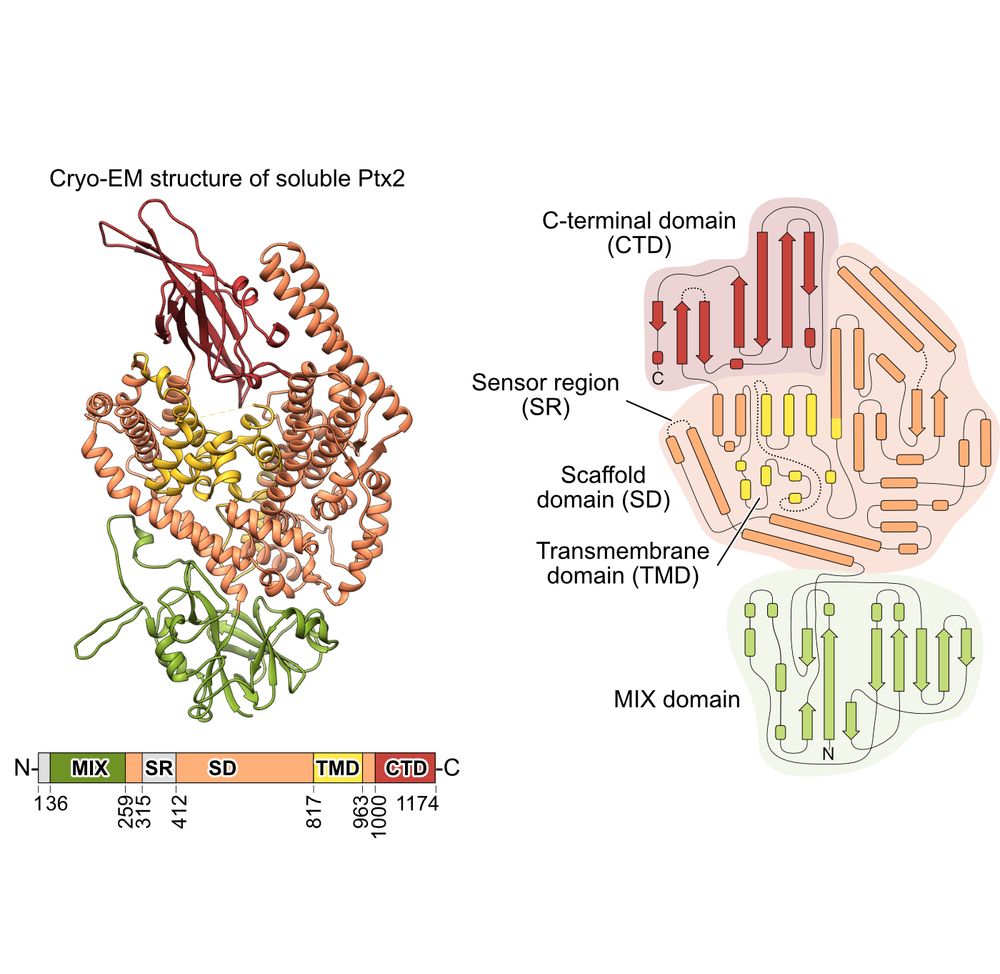
#Pseudomonas aeruginosa destroys other #bacteria using #T6SS and its effectors during infection in #cysticfibrosis patients. Here, we determined the #cryo-EM #structure of the effector Ptx2, which depolarizes target bacterial membranes.
Check it out!
www.biorxiv.org/content/10.1...
01.05.2025 15:37 — 👍 6 🔁 1 💬 1 📌 0
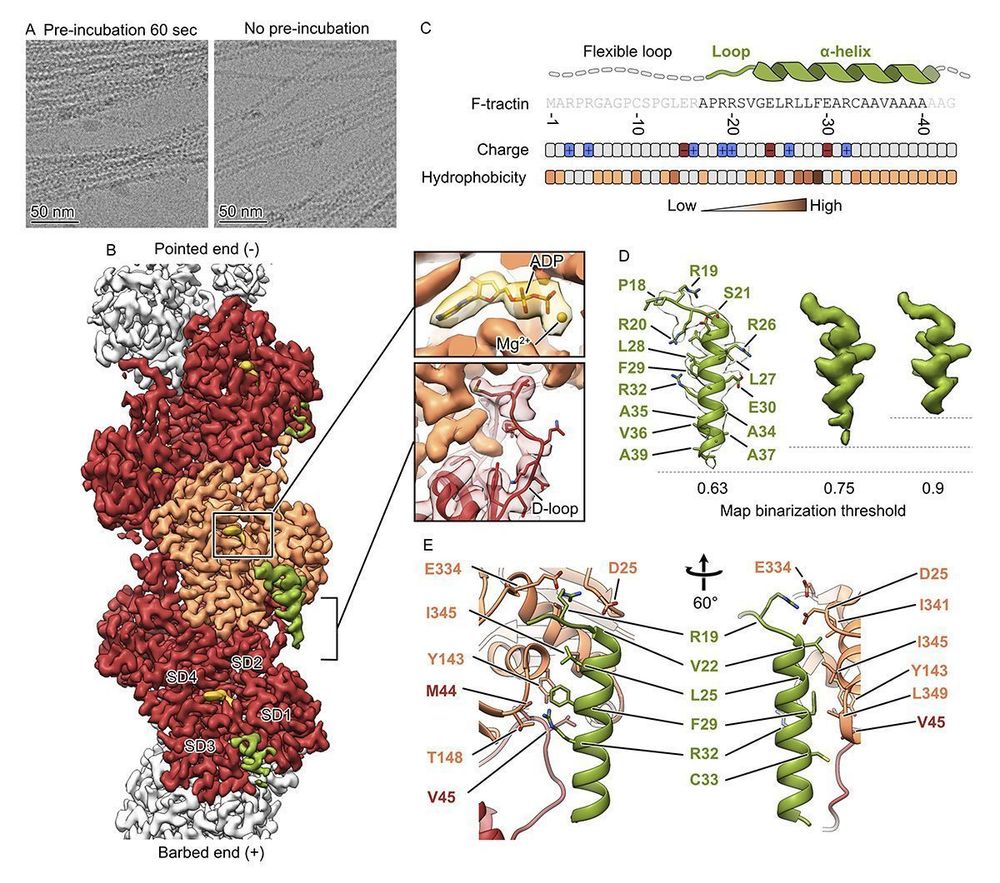
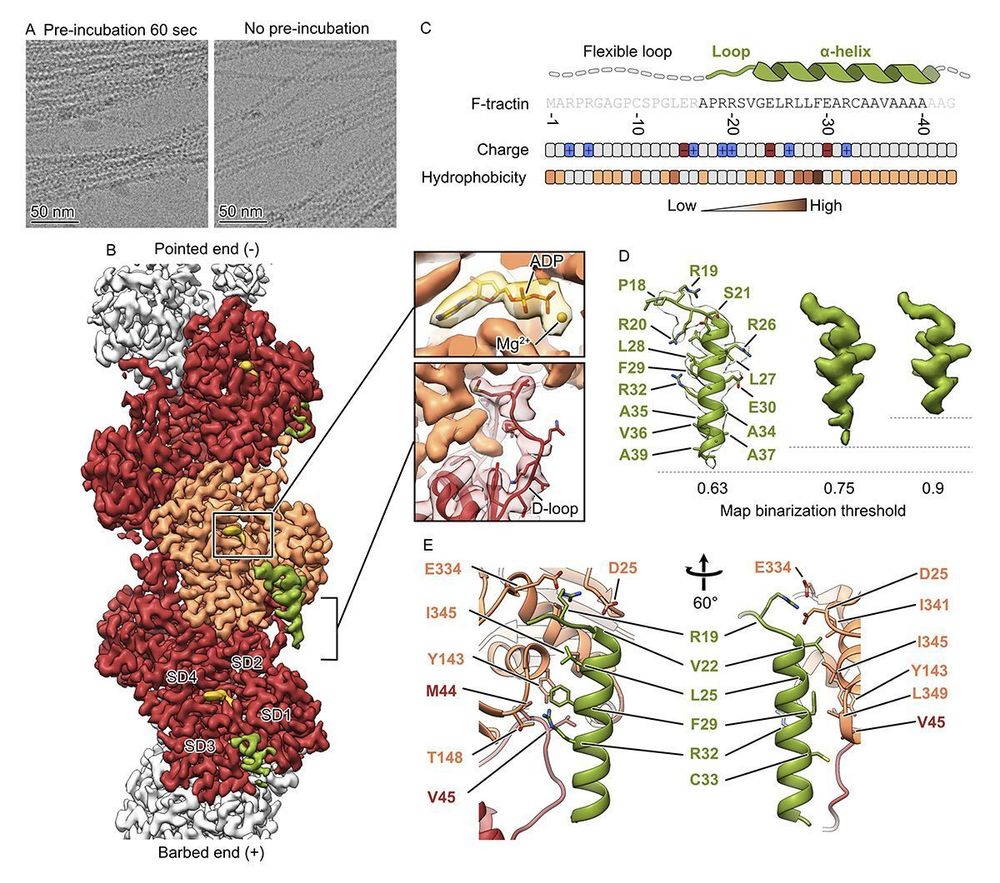
Structure of the F-tractin-F-actin complex. A new study by Dmitry Shatskiy, Athul Sivan, Roland Wedlich-Söldner @uni-muenster.de, and Alexander Belyy @alexanderbelyy.bsky.social @unigroningen.bsky.social: https://buff.ly/3EIqAur
#Cytoskeleton #StructuralBiology #Biochemistry #Actin
17.02.2025 14:07 — 👍 20 🔁 3 💬 0 📌 0
New: Using F-tractin to visualize #actin in living cells? Dmitry Shatskiy, Athul Sivan, Roland Wedlich-Söldner, and Alexander Belyy @alexanderbelyy.bsky.social determined the F-tractin-F-actin structure and showed that this probe is nearly identical to #Lifeact. https://buff.ly/3EIqAur
#cytoskeleton
10.02.2025 17:59 — 👍 21 🔁 6 💬 0 📌 0
Many thanks to @dshatskiy.bsky.social, Athul Sivan, and Roland Wedlich-Söldner for the excellent work!
#Actin #Ftractin #Cytoskeleton #Microscopy #StructuralBiology #CryoEM #Science
11.02.2025 17:38 — 👍 0 🔁 0 💬 0 📌 0
Finally, based on the structural data, we designed an optimized version of F-tractin. The result? Great labeling and minimal disturbance to actin dynamics! 🚀
11.02.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
These structural results and the biochemical data show that the previously reported differences between these peptides originate from the flexible areas of F-tractin or from differential expression levels, rather than from fundamentally different modes of interaction with F-actin.
11.02.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
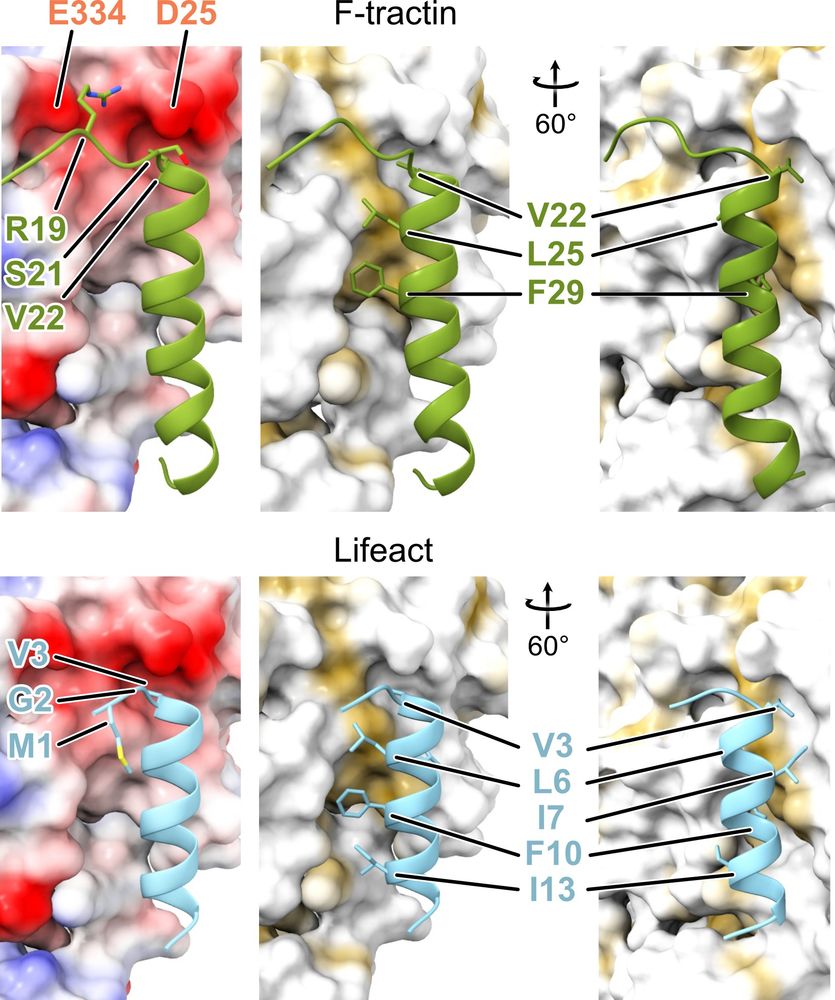
Unexpectedly, this binding site is nearly identical to the one of #Lifeact - the most popular probe for visualization of actin in living cells!
11.02.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
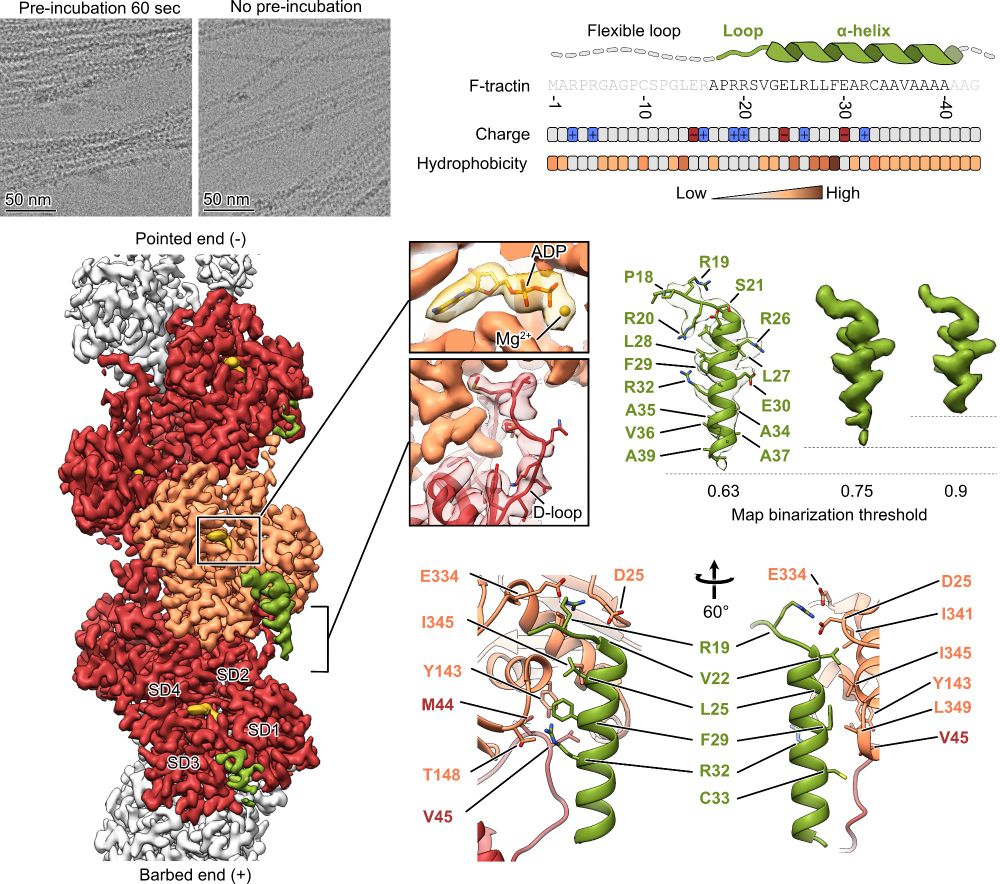
Using cryo-EM 🔬, we determined the structure of the F-tractin–F-actin complex at high resolution. We found its specific binding site, which explained its actin-binding properties!
Check out these structural insights! 📸
11.02.2025 17:38 — 👍 1 🔁 0 💬 2 📌 0
F-tractin is a small peptide derived from the rat ITPKA protein that binds F-actin. It’s a go-to tool for actin #cytoskeleton visualization in live cells. But… how does it work at the molecular level? 🤔🔍
11.02.2025 17:38 — 👍 0 🔁 0 💬 1 📌 0
Structure of the F-tractin–F-actin complex | Journal of Cell Biology | Rockefeller University Press
This study reveals the structure of F-tractin, a widely used peptide for visualizing the actin cytoskeleton, and introduces an optimized version that minim
Are you doing cell #biology? Then, let me guess, you likely stained #actin!
Our @jcb.org article reveals the structure of the popular actin probe F-tractin in complex with F-actin and introduces an optimized version for better imaging! A thread! 👀👇
rupress.org/jcb/article/...
11.02.2025 17:38 — 👍 11 🔁 2 💬 1 📌 0
If you think you know everything about enzymes, check this out, we have a surprise for you 😃
30.01.2025 19:18 — 👍 4 🔁 0 💬 0 📌 0
"Regulation of cytoskeleton dynamics" lab, at Institut Jacques Monod.
https://www.actindynamics.net/
The official social of the EMBO workshop: 1st international #Shigella meeting, being held at Institut Pasteur 20th - 24th April 2026. Abstracts and registrations are OPEN: https://www.shigella2026.conferences-pasteur.org/home
Team located on the University of Strasbourg campus (IBMC) with a remaining link with the Institut Pasteur.
Shigella, neutrophils, oxygen, RNA among other favorite research topics
Research Fellow at the University of Melbourne
enteric bacteria, evolutionary dynamics, antimicrobial resistance, genomic epidemiology
Microbial genomics - bacterial evolution, AMR, mobile elements, phylogenetics. Located at Monash University/Alfred Hospital
Lecturer in Microbiology | He/Him | Medical Microbiology | Antimicrobial Resistance (AMR) | Genomic Epidemiology | Sexually Transmissible Infections | Bioinformatics | Public Health | Views are my own | Following ≠ endorsing
Prof @University of Maryland School of Medicine Center for Vaccine Development. Shigella vaccines
I am a microbiologist, genome junky and driver of children
Postdoctoral researcher based at the University of Cambridge.
Group Leader at KCL studying host pathogen interactions
The Mikael Sellin & colleagues infection biology laboratory at Uppsala University/SciLifeLab. MS responsible for posts.
Senior Postdoc @enningalab.bsky.social, @pasteur.fr, always working on Salmonella host-pathogen interactions | Bookworm | Enjoyer of many fictional worlds | Mother of Cats | She/her 🩷💜💙
PhD student at Cambridge | AMR | genomic surveillance | microbiome
Immunologist x cell biologist x microbiologist at MRC LMB in Cambridge. Occasionally birdwatching. Less nerdy than my bio sounds. Views my own.
Prof @cambridgebiosci.bsky.social Genomic epidemiologist connecting pathogen evolution with public health outcomes. #Shigella and #AMR enthusiast. Views own. http://gen.cam.ac.uk/baker-group also www.pdu.gen.cam.ac.uk also @pducambridge.bsky.social