Great opportunity!
08.09.2025 20:00 — 👍 1 🔁 0 💬 0 📌 0@oleksiykovtun.bsky.social
Group leader @mpi_nat, Göttingen, DE, exploring mechanisms of membrane trafficking. Cryo-EM, Cryo-ET, structural biology, cell biology. All opinions are my own.
@oleksiykovtun.bsky.social
Group leader @mpi_nat, Göttingen, DE, exploring mechanisms of membrane trafficking. Cryo-EM, Cryo-ET, structural biology, cell biology. All opinions are my own.
Great opportunity!
08.09.2025 20:00 — 👍 1 🔁 0 💬 0 📌 0
First pre-print from the Lucas Lab led by Matthew Giammar and @joshdcryoem.bsky.social online today! We develop a new, extensible Python implementation of 2DTM and apply it to build pixel size optimization, constrained search and characterize molecular motions in situ www.biorxiv.org/content/10.1...
30.08.2025 00:49 — 👍 82 🔁 31 💬 8 📌 4Fascinating! Congratulations!
04.09.2025 20:01 — 👍 0 🔁 0 💬 0 📌 0
🧵In collaboration with Caroline Puente-Lelievre, Ed Moody, Rosie Cater, and Tom Williams we just submitted a new paper describing the prokaryotic origins of the COMMD protein family. @cpuentelelievre.bsky.social, @rosemaryjcater.bsky.social. @kmichie.bsky.social. 1/n
03.09.2025 00:24 — 👍 22 🔁 8 💬 2 📌 2
Delighted to share our work on cellular ubiquitination of drug-like compounds by HUWE1 - a surprising journey! Kudos to all contributors & first authors, Barbara Orth and Pavel Pohl. Sincere thanks to @ireserra.bsky.social #NatCommun for expertly guiding the winding publishing path.
rdcu.be/eDzPk
Close to magic membrane perturbation experiments to understand the ER to Golgi export.
02.09.2025 08:32 — 👍 2 🔁 0 💬 0 📌 0Building things with extensibility in mind is a true catalyst for innovation - beautifully demonstrated, I'm excited to see where 2DTM for in situ structural biology goes from here! congratulations to Matthew, @joshdcryoem.bsky.social, Laina and @bronwynalucas.bsky.social on this amazing work 🥳
30.08.2025 01:45 — 👍 7 🔁 1 💬 0 📌 0
Happy to share an exciting study from Yihong Ye’s lab at NIH, with a minor contribution from our lab: ceroid lipofuscinosis-4 (CLN4)-linked DNAJC5 mutations cause lysosomal damage as a driver of neurodegeneration in iPSC-derived neurons. CHIP safeguards lysosomes via microautophagy 👉 rdcu.be/eChof
25.08.2025 13:23 — 👍 30 🔁 12 💬 0 📌 0
One week left to apply for a fully funded PhD position in my group! Please repost. jobrxiv.org/job/max-plan...
21.08.2025 09:02 — 👍 11 🔁 4 💬 0 📌 0Textbook worth study of intracellular lipid transport.
21.08.2025 05:47 — 👍 5 🔁 0 💬 1 📌 0
Excited to share our new @pubs.acs.org paper! We engineered cells with ~10% photolipids in the ER membrane. This enabled optical control of membrane viscosity to study its impact on ER→Golgi protein transport. @dirktrauner.bsky.social @noemijimenezrojo.bsky.social
pubs.acs.org/doi/10.1021/...
It's a true superpower when things just work. Get a proper feel for it when setting up my group IT resources 😂
07.08.2025 13:56 — 👍 0 🔁 0 💬 0 📌 0Congrats Emily. Well deserved!
07.08.2025 13:48 — 👍 1 🔁 0 💬 1 📌 0Postdoc program at the MDC in Berlin, @mdc-berlin.bsky.social : take a look 👇👇
For this call, collaborative projects between two groups will be funded. Take a look at the call and the groups and drop me an email if interested. Share with colleagues too!
www.mdc-berlin.de/postdocs#t-g...

Go check out the latest preprint from Giulia Zanetti's lab. @dr-downes.bsky.social used cryo-tomography to directly visualize COPI and COPII coated vesicles in situ at unprecedented resolution in human cells. Amazingly beautiful, rigorous, insightful work.
www.biorxiv.org/content/10.1...
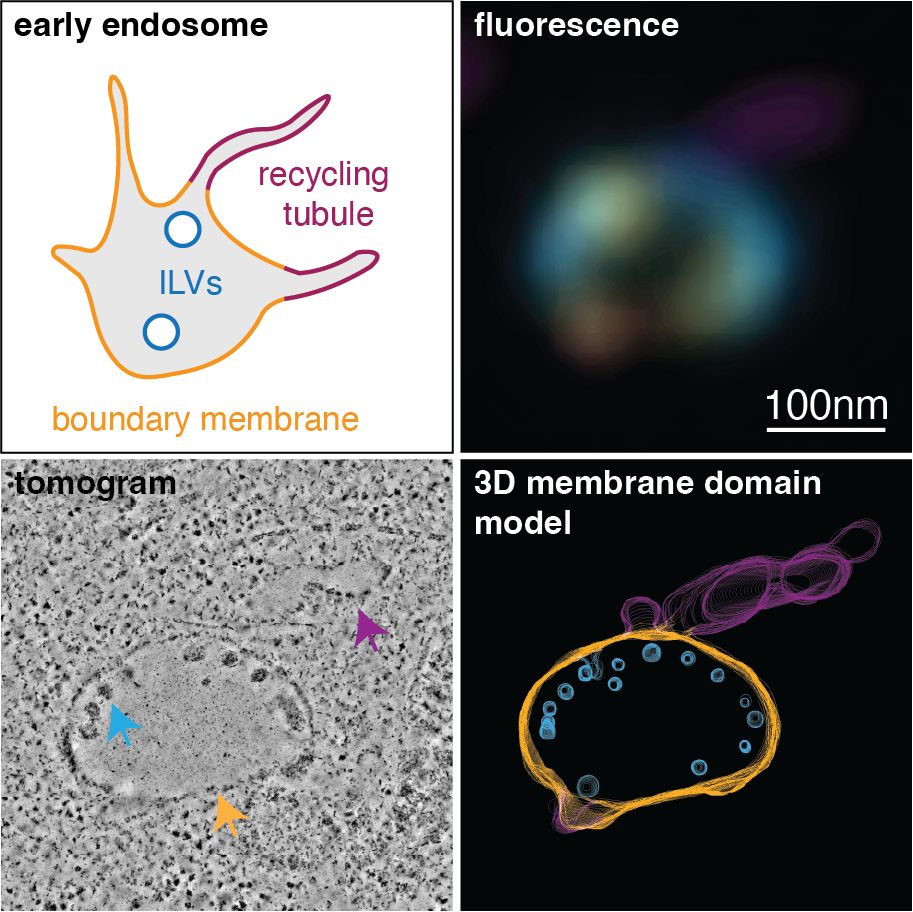
It’s incredibly hard to study lipids in biological membranes on the nanoscale. You need near-perfect information on both membrane ultrastructure and lipid density, which usually isn’t attainable. Lipid-CLEM brought to you by @mathilda95.bsky.social changes that.
www.biorxiv.org/content/10.1...
Our work on HIV-1 matrix, led by James Stacey and @dhrebik.bsky.social, is in this week's @nature.com. Extended explanatory video from @margotriggi.bsky.social.
www.nature.com/articles/s41...