
Large AI models are reported to achieve high accuracy (AUROC) predicting pathogenic variants across the genome.
A preprint reports that the predictions are based on splice variants. Using only this info (no sequences, no AI) achieves AUROC=0.944 across noncoding variants.
1/2
09.09.2025 23:01 — 👍 13 🔁 7 💬 1 📌 0

Recent advances in the inference of deep viral evolutionary history | Journal of Virology
Phylogenetic studies examining the origins, emergence, and spread of viruses have arguably been one of the most active and successful areas of evolutionary biology and form the bedrock of the flourishing field of genomic epidemiology. This, in part, reflects the ability of viruses, particularly those with RNA genomes, to evolve at rates much greater than their cellular counterparts (1). The rapid rate at which viruses evolve and accumulate mutations enables evolutionary signals to be identified through comparative genomics at short timescales relevant for outbreak investigation and response. The integration of phylogenetics and epidemiology, known as phylodynamics, has become a vital tool in response to numerous viral outbreaks, epidemics, and pandemics, including Ebola (2), Zika (3), and, more recently, COVID-19 (4) and mpox (5).
There’s been a bunch of new approaches looking at deep viral evolutionary history. We’ve put together a mini review highlighting some recent advancements in structural phylogenetics and time-dependent rate models and what they could do for the field 🦠
🔗 journals.asm.org/doi/full/10....
25.08.2025 20:32 — 👍 25 🔁 13 💬 2 📌 2

Divergent viral phosphodiesterases for immune signaling evasion
Cyclic dinucleotides (CDNs) and other short oligonucleotides play fundamental roles in immune system activation in organisms ranging from bacteria to humans. In response, viruses use phosphodiesterase...
Excited to share our new preprint co-led by @jnoms.bsky.social!
Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.
22.08.2025 19:02 — 👍 42 🔁 28 💬 2 📌 2
Thanks!
20.08.2025 20:50 — 👍 0 🔁 0 💬 0 📌 0
🙏Amazing collaboration co-led with Noor Youssef
and Navami Jain, @deboramarks.bsky.social, and our funders @cepi.net!
11/12
17.08.2025 03:42 — 👍 2 🔁 0 💬 1 📌 0
This matters for:
⚠️ Future-proof vaccine and therapeutics design
⚠️ Monitoring of high-pandemic risk viruses
⚠️ Dual-use biosecurity risk assessment
Without reliable models, we risk underestimating viral evolution—and overestimating our ability to counter it.
10/12
17.08.2025 03:42 — 👍 3 🔁 0 💬 1 📌 0

EVEREST highlights:
✅ Where models fail—and why
✅ Which viruses are least/most predictable
✅ How to estimate per-protein, model-specific reliability
✅ Concrete steps to improve ML for viral mutation prediction
9/12
17.08.2025 03:42 — 👍 6 🔁 0 💬 2 📌 0

🌍Current models fail to reliably predict mutations in more than half of the high-priority viruses identified by the WHO.
8/12
17.08.2025 03:42 — 👍 4 🔁 0 💬 1 📌 0

💪Is bigger always better? Maybe not for other taxa but for viruses - yes! For viruses, models continue to improve with increased numbers of parameters.
7/12
17.08.2025 03:42 — 👍 4 🔁 0 💬 1 📌 0

🤏Why? Viruses are severely underrepresented in training datasets (<1%) and are further downsampled after common clustering approaches.
6/12
17.08.2025 03:42 — 👍 8 🔁 0 💬 1 📌 0

📉Despite the hype, protein language models trained across the “protein universe” are outperformed by even the simplest, site-independent alignment-based model.
5/12
17.08.2025 03:42 — 👍 13 🔁 2 💬 1 📌 0
💭Imagine: It’s Day 0 of an outbreak and there’s little experiment data. Computational mutational effect predictions could provide valuable information…if we could trust them. Can we?
EVEREST doesn’t just assess performance. It also quantifies reliability for new viruses.
4/12
17.08.2025 03:42 — 👍 2 🔁 0 💬 1 📌 0
🚀To find out, we built EVEREST: Evolutionary Variant Effect prediction with Reliability ESTimation.
We benchmark models across 45 viral deep mutational scanning datasets spanning >340,000 mutations.
3/12
17.08.2025 03:42 — 👍 2 🔁 0 💬 1 📌 0
🦠 Protein language models (PLMs) have shown impressive performance in predicting mutation effects. But... viruses are a different beast.
They evolve fast, cross species, and are under pressure from host immunity. Do PLMs still work here?
2/12
17.08.2025 03:42 — 👍 3 🔁 0 💬 1 📌 0

🚨New paper 🚨
Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12
17.08.2025 03:42 — 👍 42 🔁 19 💬 3 📌 0

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
05.08.2025 06:29 — 👍 94 🔁 43 💬 1 📌 1
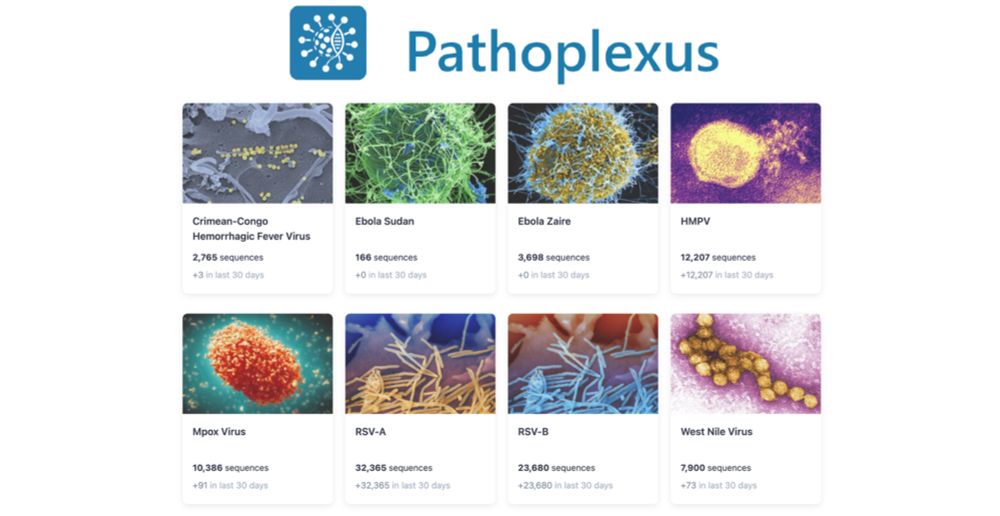
Pathoplexus | Pathoplexus July Update
Pathoplexus is a new, open-source database dedicated to the efficient sharing of human viral pathogen genomic data, fostering global collaboration and public health response.
Some great new features and updates from the awesome Pathoplexus project. This is a new open pathogen genome database that can provide access to your sequences under a use-restricted license but also feed directly in to INSDC (EBI, Genbank etc) when you are ready. pathoplexus.org/news/2025-07...
15.07.2025 14:05 — 👍 45 🔁 24 💬 1 📌 1
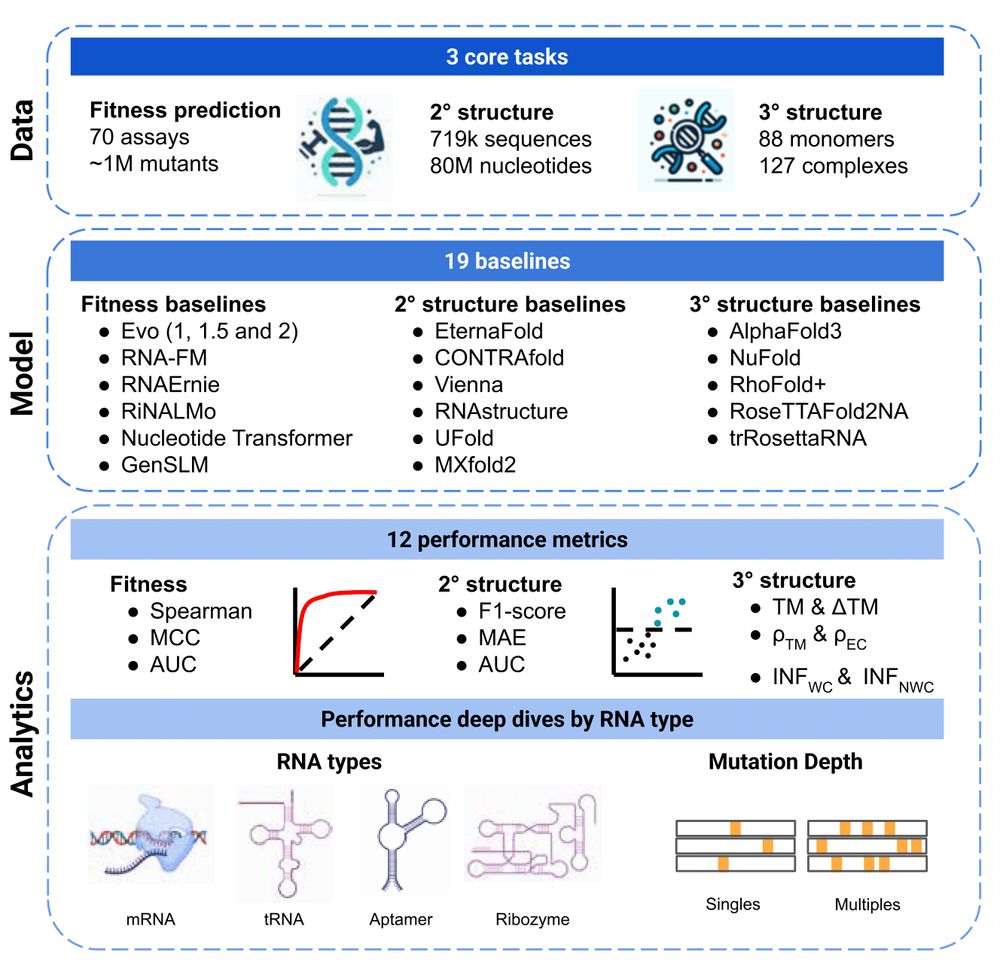
🚨 New paper 🚨 RNA modeling just got its own Gym! 🏋️ Introducing RNAGym, large-scale benchmarks for RNA fitness and structure prediction.
🧵 1/9
18.06.2025 19:35 — 👍 40 🔁 16 💬 1 📌 1
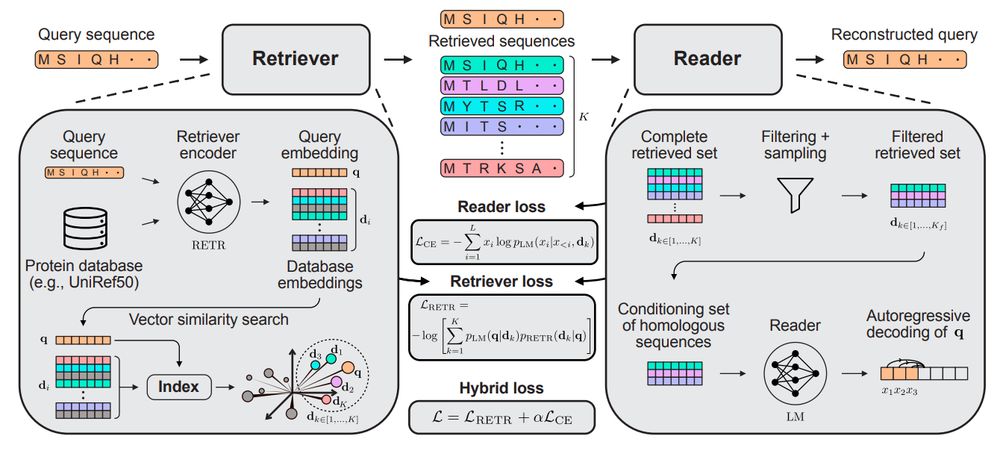
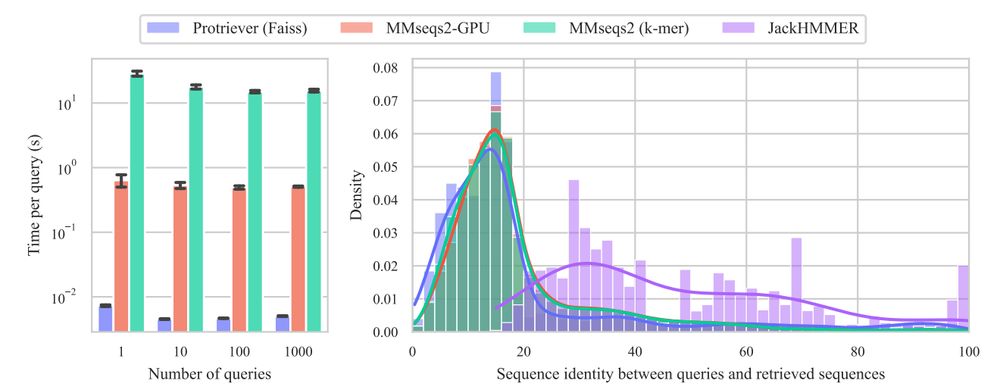
End-to-end differentiable homology search for protein fitness prediction.
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
11.06.2025 19:00 — 👍 32 🔁 9 💬 0 📌 0

Hello everyone! I am pleased to share information on the first ever Computational Structural Virology Symposium, conducted August 4th on zoom and highlighting work in this emerging field. You can register for this event here: forms.gle/CNiqskMwQEuV.... Please re-post!
12.06.2025 20:31 — 👍 68 🔁 52 💬 2 📌 6
Using genome engineering to solve humanity’s greatest problems in health, climate & sustainable agriculture. UC Berkeley, UCSF, UC Davis. https://innovativegenomics.org/
Assistant Professor UAS at HES-SO Valais-Wallis 🇨🇭 🤗 HF Fellow. Working on AI, Protein Design and Open Science. Creator of bioicons.com
European Research Infrastructure on Highly Pathogenic Agents - ERINHA AISBL
Fostering research to prevent pandemics
https://erinha.eu
PhD student (MRC-UofG CVR)
🏰 antiviral defence | 🧬 evolution | 🔮 protein structure prediction
Professor, Division of Systems Virology, The Institute of Medical Science, The University of Tokyo
Founder, The Genotype to Phenotype Japan (G2P-Japan) Consortium
Representative Director, G2P-Japan Association
Lab website: https://x.gd/1Z1lW
Tweets sometimes by Philippe Lemey, but usually by other (mysterious) lab members. Lab website: https://rega.kuleuven.be/cev/ecv
Striving to provide better evidence to improve health globally! Professor, Departments of Biostatistics, Computational Medicine and Human Genetics, UCLA
Assistant Professor @ Stanford Genetics & BASE Initiative. Mapping the regulatory code of the human genome to understand heart development and disease. www.engreitzlab.org
Scientific Program Leader @ Gladstone Institute of Virology with Melanie Ott. Previously (and always), an imaging aficionado!
Resident panhandler, http://trichelab.org/
Posts may contain trace quantities of blood🩸, chromatin 🧬, and stats 🧮
CV: https://scholar.google.com/citations?user=AOoIO74AAAAJ
Views expressed are my own (but for the right price they can be yours!)
Duke University alumn. Virology research specialist in the Sheahan Lab at CVRG (y'know, at that other blue school). Pop punk millennial. she/they. ✡️•🏳️🌈•🖖
All things microbiota for now but a generalist in computation and genomics
2-3 sketches a day keeps it away
All posts and opinions are mine and mine only
Professor Emeritus (University of Manchester), writes on biology, history of science & French Resistance. Biography of Francis Crick, out in Nov 2025.
Link to publications: https://orcid.org/0000-0002-8258-4913
Virology lecturer at Nantes University. Interested in polyomaviruses, particularly capsid interactions with antibodies and host receptors.
My scientific passions: microbiomes and bioinformatics
Find me also on LinkedIn: https://www.linkedin.com/in/cedric-laczny-02b720150/
or ORCID: https://orcid.org/0000-0002-1100-1282
Physician-Scientist
Hematopathologist
Medsky
Scientist, Infectious Disease Modeler, Views are mine
computational protein scientist
vmap enthusiast
🧬👩💻🧬
github.com/justktln2
Infectious diseases, microbiology, virology, public health and particularly emerging infections & genomics @UKHSA


















