Seeds for the different Kronos SPL-resistant lines(rSPLs) that accelerate wheat heading time are available without restrictions from GRIN-GLOBAL under accession numbers PI 707858 (rSPL-A3), PI707859 (rSPL-A4), PI 707860 (rSPL-A13), PI 707861 (rSPL-B3), and PI 707862 (rSPL-B13)
12.01.2026 19:11 — 👍 1 🔁 0 💬 0 📌 0

Our paper integrating the endogenous age pathway with the vernalization, photoperiod and GA pathways in the regulation of heading time in wheat is now published in The Plant Journal (open access). onlinelibrary.wiley.com/doi/epdf/10....
12.01.2026 19:11 — 👍 2 🔁 0 💬 1 📌 0
Finally, we developed dominant miR156-resistant alleles rSPL3, rSPL4, and rSPL13 that accelerate wheat heading time that can be used to improve wheat adaptation to changing environments.
08.11.2025 20:46 — 👍 0 🔁 0 💬 0 📌 0
Additionally, We showed that DELLA proteins can interact with SPL proteins reducing their ability to induce flowering and that interactions between DELLA and both VRN1 and FUL2 can compete with the DELLA-SPL interactions, likely reducing DELLA’s ability to repress SPL3 and SPL4 activity.
08.11.2025 20:46 — 👍 0 🔁 0 💬 1 📌 0
Earlier heading was associated with the upregulation of miR172 and flowering-promoting genes VRN1, FUL2, and FT1 and the downregulation of flowering-repressing genes AP2L1 and VRN2.
08.11.2025 20:46 — 👍 0 🔁 0 💬 1 📌 0
We also characterized the interactions between the age, photoperiod, vernalization and GA pathways in pasta wheat. Plants with reduced levels of microRNA156 or higher expression of SPL3, SPL4, and SPL13 exhibited accelerated heading time, with stronger effects under suboptimal inductive conditions.
08.11.2025 20:46 — 👍 0 🔁 0 💬 1 📌 0

Our manuscript on the role of the SPL genes and the endogenous age pathway on wheat heading time is now available in bioRxiv!
www.biorxiv.org/cgi/content/...
08.11.2025 20:46 — 👍 2 🔁 0 💬 1 📌 0
Wheat spike imputed Resolve simFISH | Dubcovsky Lab
Explore the spatial distribution of the 74,464 genes imputed from the single cell RNA-seq into the spatial transcriptomics at dubcovskylab.ucdavis.edu/imputed-genes.
13.10.2025 19:04 — 👍 3 🔁 0 💬 0 📌 0
JD99 wheat spike smFISH spatial transcriptomics | Dubcovsky Lab
You can visualize the spatial distribution of the 99 genes included in the spatial transcriptomics analysis at dubcovskylab.ucdavis.edu/JD99-wheat-s...
13.10.2025 19:04 — 👍 3 🔁 0 💬 1 📌 0

Our Genome Biology paper on wheat spike development integrating single cell and spatial transcriptomics is now published! Check out the paper here: doi.org/10.1186/s130... and enjoy the beautiful pictures (including our 44 supplementary figures)! Be sure to check out some of our tools below!
13.10.2025 19:04 — 👍 41 🔁 20 💬 2 📌 2
Wheat spike imputed Resolve simFISH | Dubcovsky Lab
74,464 genes with imputed expression from single cell RNA-seq into spatial transcriptomics cells dubcovskylab.ucdavis.edu/imputed-genes.
13.08.2025 18:31 — 👍 4 🔁 1 💬 0 📌 0
We also developed two WEB tools to visualize gene expression in cells of wheat developing spikes linked in the thread below!
13.08.2025 18:31 — 👍 4 🔁 1 💬 1 📌 0

Spatial and single-cell expression analyses reveal complex expression domains in early wheat spike development
Wheat is important for global food security and understanding the molecular mechanisms driving spike and spikelet development can inform the development of more productive varieties. In this study, we...
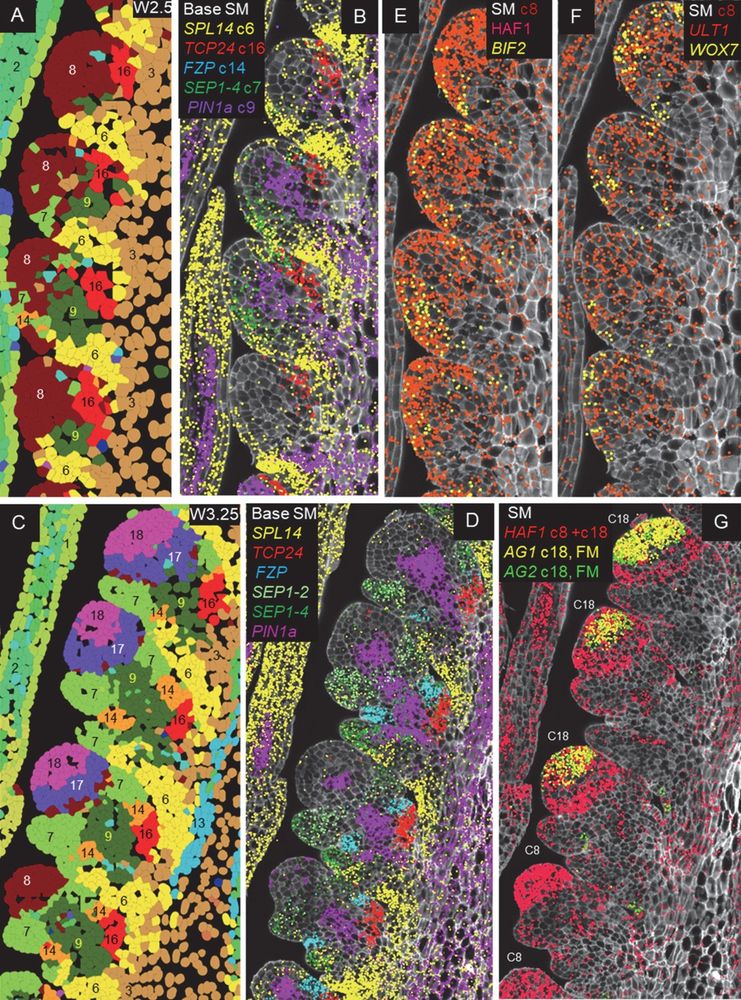
An update to our spatial transcriptomics study of the wheat spike is now available in bioRxiv! The updated version includes functional validation of the roles associated with the specific expression patterns of LFY in intercalary meristems, SPL14 in inflorescence meristems, and FZP in glume axillae.
13.08.2025 18:31 — 👍 22 🔁 6 💬 1 📌 0
In this paper, we report the transfer of the stripe rust resistance gene Yr78 to durum wheat (PI 702944) and the combination of the resistance genes Yr36 and Yr78 in coupling in common wheat (PI 706442) to facilitate their simultaneous deployment in wheat breeding programs.
10.08.2025 17:15 — 👍 1 🔁 0 💬 0 📌 0
Stand Up for UC
We must stand together to protect our students, staff, faculty and our mission.
The federal government's $1B dollar demand from the University of California is designed to devastate public Universities and their critical mission. Our lab will #StandUpForUC to defend one of America’s greatest public university systems and support this life-saving work. ucal.us/standupforuc
09.08.2025 12:52 — 👍 7 🔁 0 💬 0 📌 0

Talking Biotech 467 - #Celiac safe wheat? Less immunoreactive wheat has been developed by
@UCDavisPlants using old-school mutagenesis. A scintillating talk with grad student Maria Rottersman.
@ASPB
share.transistor.fm/s/9419052e
12.07.2025 05:44 — 👍 17 🔁 8 💬 2 📌 1
Old Tricks, New Wheat for Celiacs - Maria Rottersman
SummaryIn this episode of the Talking Biotech Podcast, Kevin Folta...
🎙️ Just published a new episode of Talking Biotech with Dr. Kevin Folta: Old Tricks, New Wheat for Celiacs - Maria Rottersman. Have a listen:
12.07.2025 05:00 — 👍 9 🔁 3 💬 0 📌 0
Deletion of wheat alpha-gliadins from chromosome 6D improves gluten strength and reduces immunodominant celiac disease epitopes
New work from a graduate student, Maria Rottersman, and others in the lab demonstrates that deleting the alpha-gliadins on chromosome 6D of wheat improves gluten strength and produces less celiac disease epitopes. Check it out now in Theoretical and Applied Genetics: rdcu.be/egXZ2
09.04.2025 04:18 — 👍 10 🔁 1 💬 0 📌 1
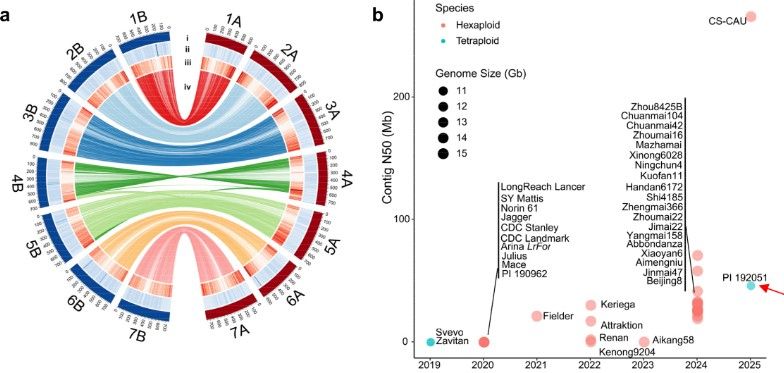
A high-quality genome assembly of the durum wheat landrace PI 192051. a Circular diagram showing the assembly features of PI 192051. The tracks, arranged from outermost to innermost, include: (i) chromosomes, (ii) GC content, (iii) gene density, and (iv) syntenic connections between A and B subgenomes. b Comparison of genome assembly quality (contig N50) between PI 192051and other published tetraploid (turquoise blue) and hexaploid (pink) wheat genomes. The horizontal axis represents the year, and the vertical axis represents the contig N50 size. Each circle represents one genome, with the diameter of the circle corresponding to the genome size. PI 192051 is highlighted with a red arrow. c Syntenic blocks among T. urartu (G1812 v2.0), T. monococcum (PI 306540), T. durum (PI 192051 and Svevo v1.0), T. dicoccoides (Zavitan v1.0), Ae. searsii (TE01), Ae. speltoides (TS01), and the A/B subgenomes of the bread wheat variety Chinese Spring (CS, RefSeq v2.1). Each line represents a syntenic block of 15 or more gene pairs with ≥ 80% identity.
🌱From a Nature Portfolio preprint: Cloning of Lr30 reveals it’s identical to Lr.ace-4A and encodes a unique NLR gene conferring strong rust resistance in durum #wheat, but less so in hexaploid. (Jorge Dubcovsky)
▶️ www.researchsquare.com/article/rs-6...
#PlantScience #PlantImmunity
01.04.2025 15:46 — 👍 9 🔁 2 💬 1 📌 0
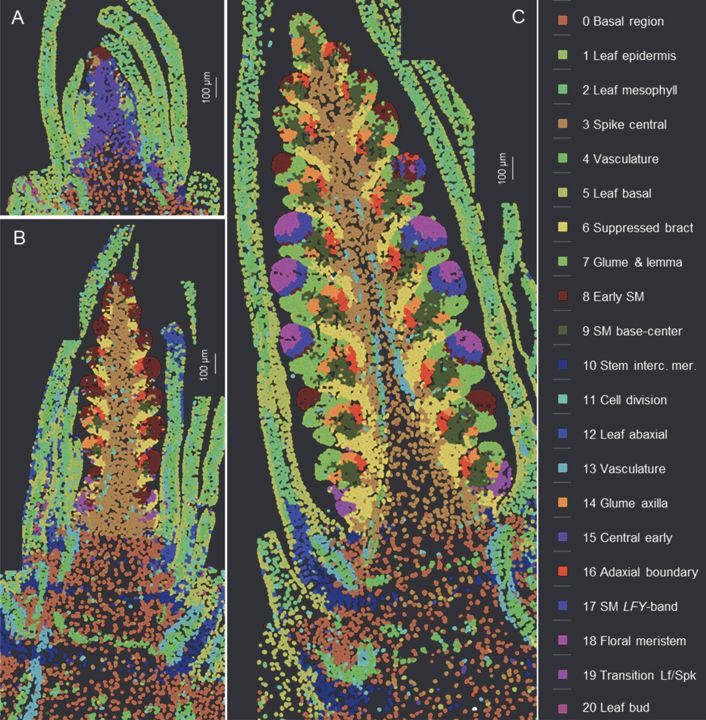
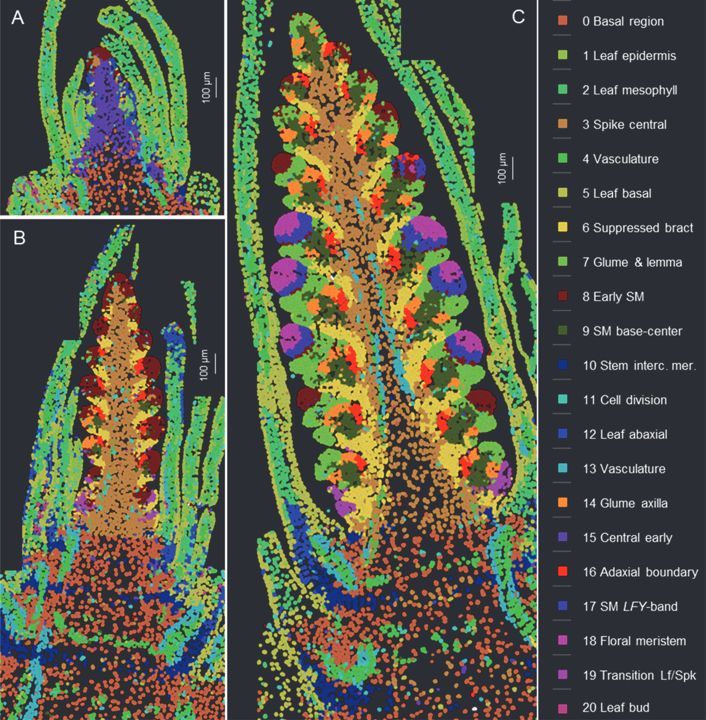
Three stages of wheat spike development with color-coded labels showing gene expression clusters. Several structures are visible clearly changing in form and gene expression pattern between the three developmental stages.
#PlantSci Research Weekly March 7 plantae.org/plant-scienc... (2/2) Single-cell analysis of wheat spikelet; Autoactivated calcium channel enhances symbiosis; Fungal pathogen hijacks phosphate signaling; Erucamide inhibits the type III secretion system; How Rhodanobacter R179 evades plant immunity
07.03.2025 07:31 — 👍 11 🔁 5 💬 1 📌 1
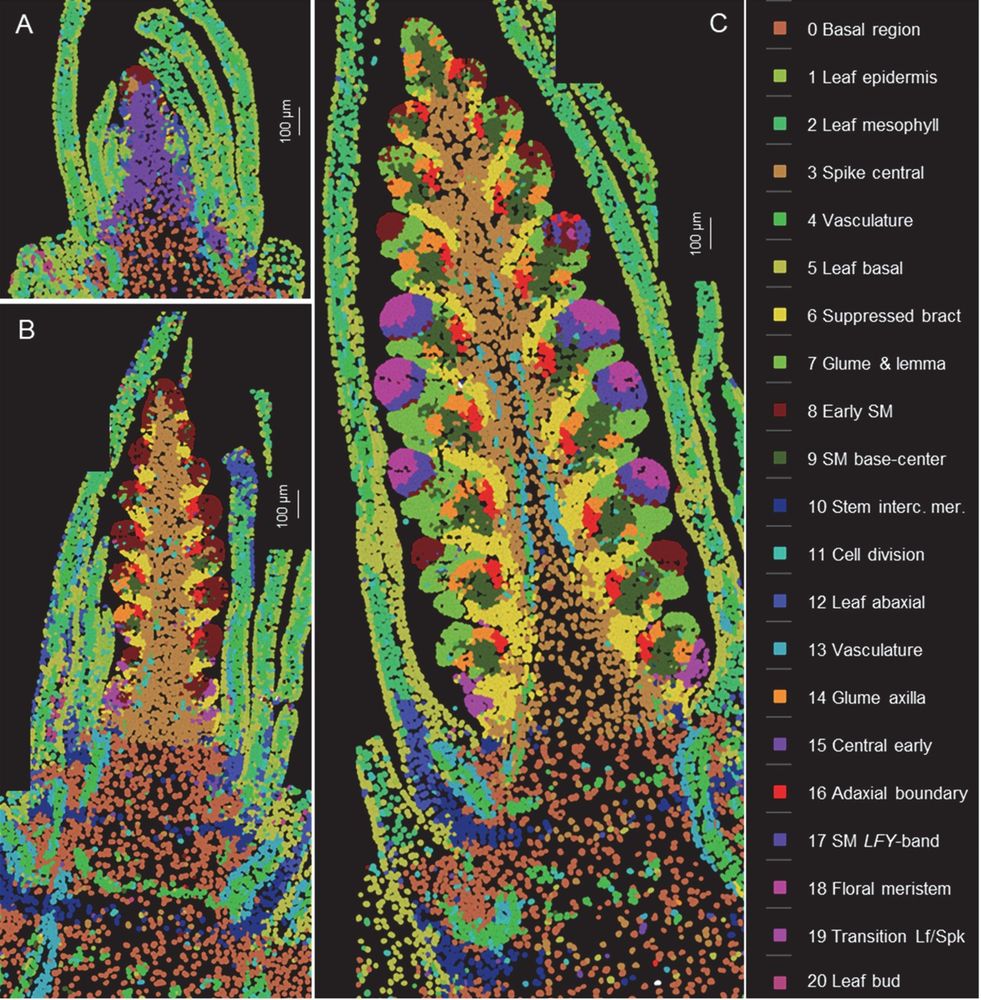
Spatial cell clusters based on the expression of 99 genes analyzed by smFISH at three stages of wheat spike development.
Expression profiles of genes differentially expressed during spikelet development.
We’re excited to share our new preprint on wheat spike development! We used spatial transcriptomics and scRNA-seq to take a closer look at the different cell types and expression domains during key developmental stages of the wheat spike. Check it out here:
www.biorxiv.org/content/10.1...
20.02.2025 17:39 — 👍 59 🔁 25 💬 0 📌 5
Integrative Postharvest Biologist @ucdavisplants. | 🍅🥔 Metabolism & Physiology|Compassionate Excellence| ♥️ Barbados 🇧🇧|♥️ Sea turtle conservation 🐢|
UC Davis rice geneticist & co-author of Tomorrow's Table: Organic Farming Genetics & the Future of Food. TED talk. My lab studies genetics for crop resilience & #plant-microbe interactions. Volunteer Ranger USFS. Nature photography
Wheat geneticist and group leader at John Innes Centre. Interested in #micronutrients, #senescence, #polyploidy and #genomics. She/her. https://borrilllab.com/
Assistant Professor, Univ of Pennsylvania. Plant developmental biology, robustness, evolution, transcription factors, Bajan rum, LFC
Plant Biologist @CNRS: Flowers 🌼, Plant Development, Gene Regulation, Evolution and Rock climbing 🧗♂️🚴♂️🏔️⛷️ Science & Society L'histoire secrète des fleurs + Les clés du champ @humensciences
Biologist of plants and microorganisms, Prof at #UCDavis
🍇🧬 Everything Vitis and associates
https://cantulab.github.io
https://www.grapegenomics.com
Professor & Assoc Dept Head UofA EEB (@uofa-eeb.bsky.social) in Tucson studying plant evolution, botany, polyploidy, chromosomes, and biodiversity. Views are my own. He/Him https://www.barkerlab.net
Plant developmental biologist - flowering, inflorescence architecture and heat responses in wheat, barley and oats 🌾.
University of Adelaide, Australia.
Professor at UC Davis, studying plant immunity and bacterial pathogens. Loves weightlifting, running, and the outdoors. Views are my own.
Promoting SciComm, teaching, equity & inclusion in plant biology. #PlantScience
PI NSF RCN: ROOT&SHOOT. https://rootandshoot.org/
Features Editor ThePlantCell, PlantPhysiology
@PlantTeaching@genomic.social
https://linktr.ee/PlantTeaching 🏴
Research group focused on Evo-Devo of climbing plants 🌿
We are part of the Department of Botany at UFRGS.
Associate Professor UC Berkeley, puzzle-solver interested in comparative immunology 🌱🍄🟫🧬 and computational biology 💻
Dr Alexander J. Hetherington | Plant evolutionary biologist, University of Edinburgh UK | UKRI Future Leader Fellow |
Lab website: https://www.ed.ac.uk/biology/groups/hetherington
Plant biologist at UCSD (how auxin regulates transcription across land plants, from receptors to pol II and the steps in between). Mostly, moss⬌Arabidopsis. Grew up in Minnesota, attended/worked at UMinn, UO, UM, and IU.
Prof. Plant Adaptation UniPotsdam, head of Dept Leibniz IGZ. Interested in how plants adapt to climate change.
Or just “Li” |
Assist. Prof. @ Plant Bio Michigan State U. |
Also post data visualization |
Lab: https://cxli233.github.io/cxLi_lab/ |
GitHub: https://github.com/cxli233
Associate Professor and Associate Nematologist at UC Davis. Parasites, Plants, Nutrients, Immunity 🪱🌱🍅💉
www.nemaplant.org
Manager UC Davis Plant Transformation Facility
🌱 Discussion of all plant-related research and concepts.
UC Davis supports academic and research programs in plant sciences, plant biology, plant pathology, viticulture and agricultural engineering.
https://instagram.com/ucdavisplants