
New Title Alert: ARCTIC-3D- a software for data-mining and clustering of protein interface information.
Learn more here: buff.ly/IbbwP2N
#SBGridSoftware #StructuralBiology #SBGrid
@jpglmrodrigues.bsky.social
Computational Biologist and Team Lead at Schrodinger. Proteins & Python. Opinions are my own. https://joaor.eu

New Title Alert: ARCTIC-3D- a software for data-mining and clustering of protein interface information.
Learn more here: buff.ly/IbbwP2N
#SBGridSoftware #StructuralBiology #SBGrid

From @jdrakephd.bsky.social & @bahanbug.bsky.social in @plos.org #Computational #Biology | How to write a scientific paper in fifteen steps | #Education #Science 🧪 🔓 #GoodReadingForAll CC/ @ppalagi.bsky.social
⬇️
journals.plos.org/ploscompbiol...

The next Bay Area COMP Together is Thu 2 Oct, featuring Jit Mukherjee (Gilead) describing the discovery of lencapavir and Jeff Blaney (Genentech) on lessons learned in 42 years of drug discovery!
www.linkedin.com/posts/comp-t...
🚨 Super excited for our new preprint on flexibility cues in AlphaFold!
Together with @ezgikaraca.bsky.social and @aysebercinb.bsky.social, we found that distograms of AF2.3 and AF3 mirror MD sampling by predicting the extent of a novel conformational change! 🤯
For more👇
We just bumped into something very preliminary… but very exciting:
AF2.3 and AF3.0 distograms may potentially reproduce MD-like behavior.
Until we do further tests, you can reach our early insights at www.biorxiv.org/content/10.1...

A small reminder to all structural biologists around working on biomolecular complexes: please consider sharing your complexes as targets for CAPRI - AI has not solved all structure prediction problems and there are still challenges! See www.capri-docking.org/contribute/
29.07.2025 06:26 — 👍 27 🔁 16 💬 2 📌 1Exploring the Potential of AlphaFold Distograms for Flexibility Assignment in Cryo-EM Maps www.biorxiv.org/content/10.1101/2025.07.25.666757v1 #cryoEM
28.07.2025 18:40 — 👍 2 🔁 1 💬 0 📌 0
A paper thar took too long to finish. We show that AlphaFold3 its clearly better than AlphaFold2 (and Boltz-1 and Chai-1) to predict the structure of antibody-Antigens, but only for cases with (har to detect) similarity in the training set.
www.biorxiv.org/content/10.1...
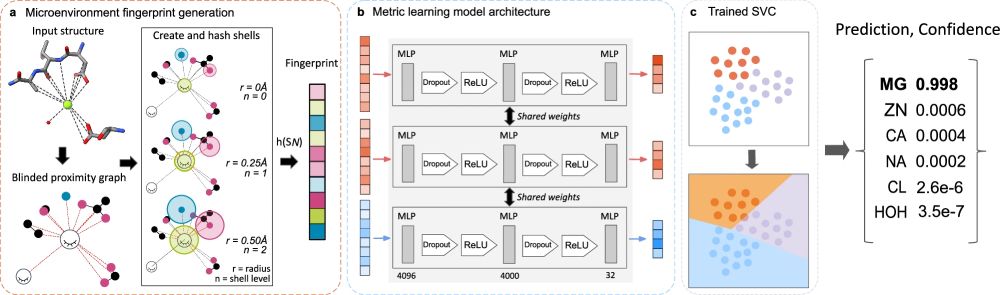
Happy to share that our paper on Metric Ion Classification (MIC) is now published!
www.nature.com/articles/s41...

Calling all structural biologists in the NYC metro area! The nysbdg.bsky.social is back for in-person events!
Register here 👉🏼 buff.ly/dGmJeau
I look forward to seeing you there! 😎
#structuralbiology #cyrstallography #cryoEM #NMR
#chromatography #proteinpurification #AKTAsystems

Interested in a postdoc position combining computational protein design and molecular simulation? There is an open position in my group at the MRC-LMB in Cambridge, UK.
www.nature.com/naturecareer...
Feel free to message me with any questions.
With contributions from fantastic colleagues @martinsteinegger.bsky.social , @mikeinouye.bsky.social, @jlistgarten.bsky.social , @ideasbyjin.bsky.social, @michael-heinzinger.bsky.social, and many more, the first CSHL volume on ML for Protein Science and Engineering is out: lnkd.in/dQdgGPpp
15.06.2025 11:30 — 👍 15 🔁 4 💬 1 📌 2
Only a few days left to apply for our EMBO course with a great lineup of speakers and tutors! Please feel free to RT!
Course website: meetings.embo.org/event/25-bio...
Organizers: myself, @amjjbonvin.bsky.social @bioinfo.se @lindorfflarsen.bsky.social
Location: www.ibg.edu.tr
To put it simply, Europe funds education, the US funds research.
11.06.2025 01:13 — 👍 1 🔁 0 💬 1 📌 0I get it. I was trying to make the point that Europe simply cannot make up for those cuts because it's a fundamentally different system.. in my opinion, making any type of comparisons is pointless.
11.06.2025 01:11 — 👍 0 🔁 0 💬 1 📌 0And yet ~50% of postdocs in the US are foreigners :) I wouldn't compare different systems, built on societies with different sets of values. If Europe (et al) invests more as a result of US divestment, that's a net positive: more diversity of ideas, less concentration of IP, the list goes on..
11.06.2025 00:07 — 👍 0 🔁 0 💬 1 📌 0New paper out www.frontiersin.org/journals/bio...
VTX is available at github.com/VTX-Molecula...
@cg-martini.bsky.social @janstevens.bsky.social
@maximemaria.bsky.social @sguionni.bsky.social @pltc.bsky.social @qubit-pharma.bsky.social
The rendering style got me half expecting to see Goku and Vegeta pop up at any moment from behind a helix! Beautiful! Great job!!
10.06.2025 15:04 — 👍 0 🔁 0 💬 1 📌 0This paper combines two sort-of-crazy (in the best sense) approaches in cryoEM sample preparation:
(1) Native, soft-landing electrospray ion beam deposition, followed by (2) deposition of amorphous ice, and reheating and freezing by laser flash melting. Has potential for time-resolved studies.
Depending on where you'll be, try and look for local cycling/wheelers clubs. They're usually nice, organize rides, etc. A quick search for Boston shows www.crw.org which actually seems really nice!
09.06.2025 16:25 — 👍 1 🔁 0 💬 0 📌 0Ah! Tomayto, tomahto 😜
08.06.2025 03:26 — 👍 1 🔁 0 💬 0 📌 0Really cool work on diffusing structures into cryo-EM densities by Ellen Zhong et. al arxiv.org/html/2506.04...
#cryoem
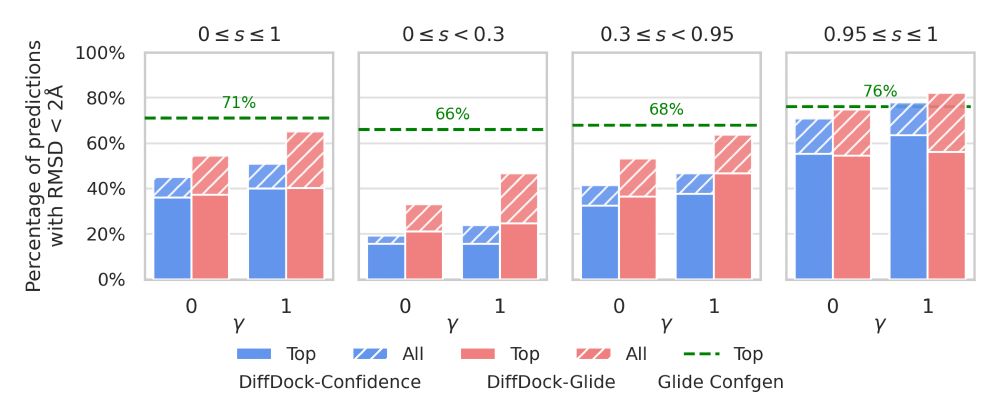
Several bar plots showing the improved performance and ability to generalize to unseen targets of Diffdock-Glide vs standard Diffdock.
New preprint from my colleagues at Schrödinger on blending ML methods with physics-based refinement and scoring to improve small molecule docking performance - and generalization - on several tasks for early stages of drug discovery. Link👇
www.biorxiv.org/content/10.1...
#chemsky

The manuscript describing the new modular version of HADDOCK is finally available as a preprint! The result of a team work over several years, supported by @bioexcelcoe.bsky.social @bijvoet-centre.bsky.social @esciencecenter.bsky.social - www.biorxiv.org/content/10.1...
07.05.2025 15:50 — 👍 22 🔁 13 💬 1 📌 0This has been a long journey, but it's now out in final form. @kjamali.bsky.social
academic.oup.com/bioinformati...
Anybody know on TED (The Encyclopedia of Domains), what the domain-domain "Interaction score" is? I can't find it in the paper on or on the website. Example: kinase N and C terminal domains for EGFR have a score of 7.0:
ted.cathdb.info/uniprot/P00533
You can send me an email at firstname.lastname@schrodinger.com, or DM me here, or LinkedIn, whatever you prefer!
16.03.2025 06:56 — 👍 3 🔁 0 💬 1 📌 0
My team at Schrödinger is looking for 2 interns to join us in NYC for 3 months this summer!
If you are comfortable writing Python, know (even a little!) about protein structure, and can work in the US, feel free to reach out!
Check the kind of work we do 👇
tinyurl.com/43j57uww

Google Summer of Code projects are available this year to improve molecular simulation in Molly.jl.
Feel free to ask me any questions.
julialang.org/jsoc
Many thanks to @sbgrid.bsky.social for inviting us to talk about our structure refinement tools for #cryoem - better structures enable better predictions. It was a pleasure to share the work of many talented group members over the years!
youtu.be/g11qEvSmTLs