
Dr. Jane Goodall, 1934-2025
An exceptional scientist, and a singular woman in science
#WomeninScience 🐒
www.nytimes.com/2025/10/01/o...
@mattetrovato.bsky.social
Passionate about (epi)genomics and chromatin biology. Postdoc at @ifomresearch.bsky.social (Milan 🇮🇹) @LabPagani // @AIRC_it & @MSCActions Fellow // Alumnus @embl.org (Heidelberg 🇩🇪) 🔬🧬🦕 📣 Latest: tinyurl.com/H33K9AK27A

Dr. Jane Goodall, 1934-2025
An exceptional scientist, and a singular woman in science
#WomeninScience 🐒
www.nytimes.com/2025/10/01/o...
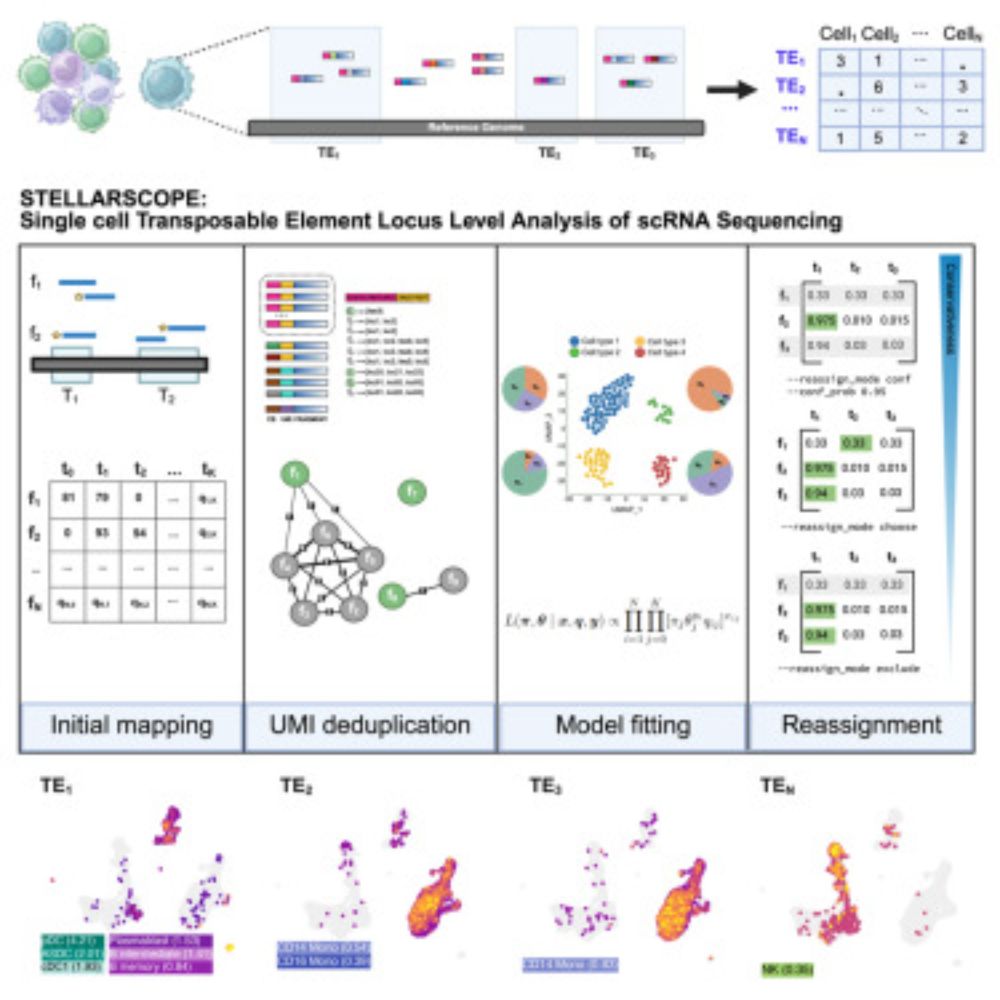
🧵1/ Introducing Stellarscope: an EM-based tool for quantifying locus-specific transposable element expression in single cells.
A single-cell transposable element atlas of human cell identity: Cell Reports Methods www.cell.com/cell-reports...
@hamarillo.bsky.social #TEHub #singlecell #genomics
What a great week at @earlhaminst.bsky.social, spent together with the awesome @longtrec.bsky.social folks! Learned a lot and had some great discussions about long-reads sequencing analysis! 🙌🏻💡
Big thanks to @anaconesa.bsky.social and everyone who made it happen! 👏🏼
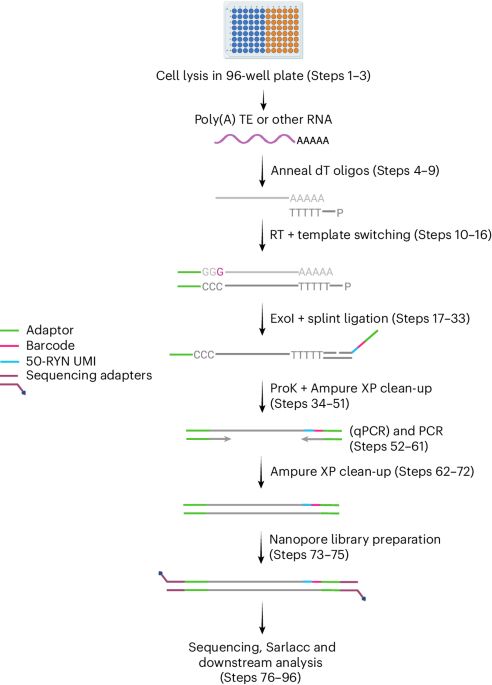
Very happy to share our protocols paper for CELLO-seq. This will make single cell long read RNA-seq more accessible and provides analysis guidelines. We hope this helps the #transposon #TEsky community and folks working on #singleCell isoform and allelic #gene expression. doi.org/10.1038/s415...
16.07.2025 16:55 — 👍 104 🔁 38 💬 8 📌 1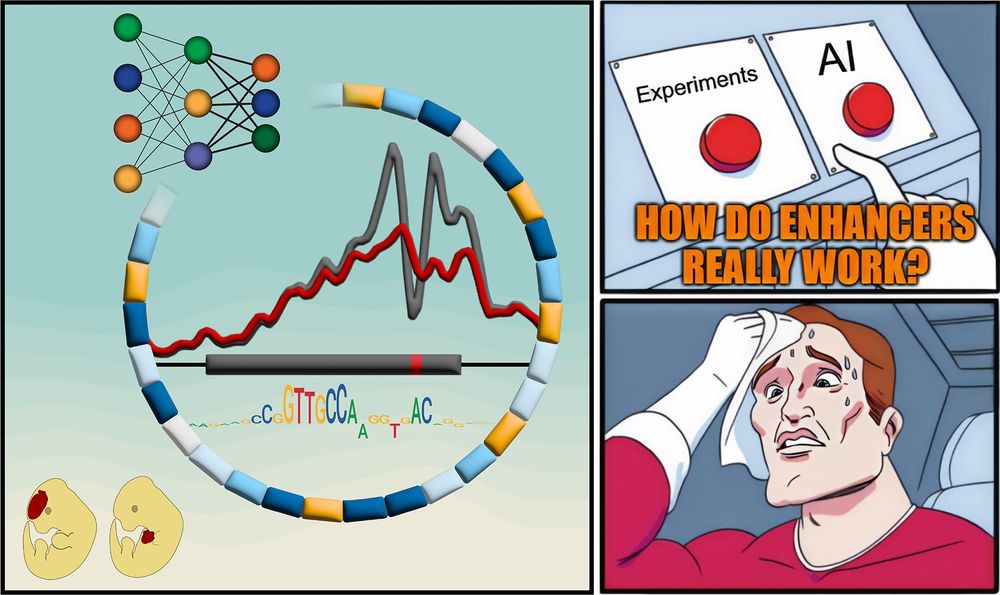
A meme-style comic panel with three parts. Left: A stylized enhancer with a mutation, surrounded by colored blocks representing functional motifs, a neural network diagram, chromatin accessibility signal traces, and a sequence motif. Two cartoon mouse embryos below show different LacZ reporter activity patterns. Top right: A hand hovers anxiously between two red buttons labeled “Experiments” and “AI,” with the caption “HOW DO ENHANCERS REALLY WORK?” Bottom right: A sweating superhero wipes his forehead, looking stressed about the difficult choice.
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22

Delighted & excited to share our latest preprint on non-canonical enhancers in human pluripotent stem cells, the result of a fantastic and fun collaboration with @guenesdoganlab.bsky.social:
www.biorxiv.org/content/10.1...
🧵 below
If you're at #ESHG2025, don't miss the chance to visit the poster and have a chat with my friend @sanchezgaya.bsky.social!!! 👏🏼😎
25.05.2025 12:47 — 👍 5 🔁 1 💬 0 📌 0
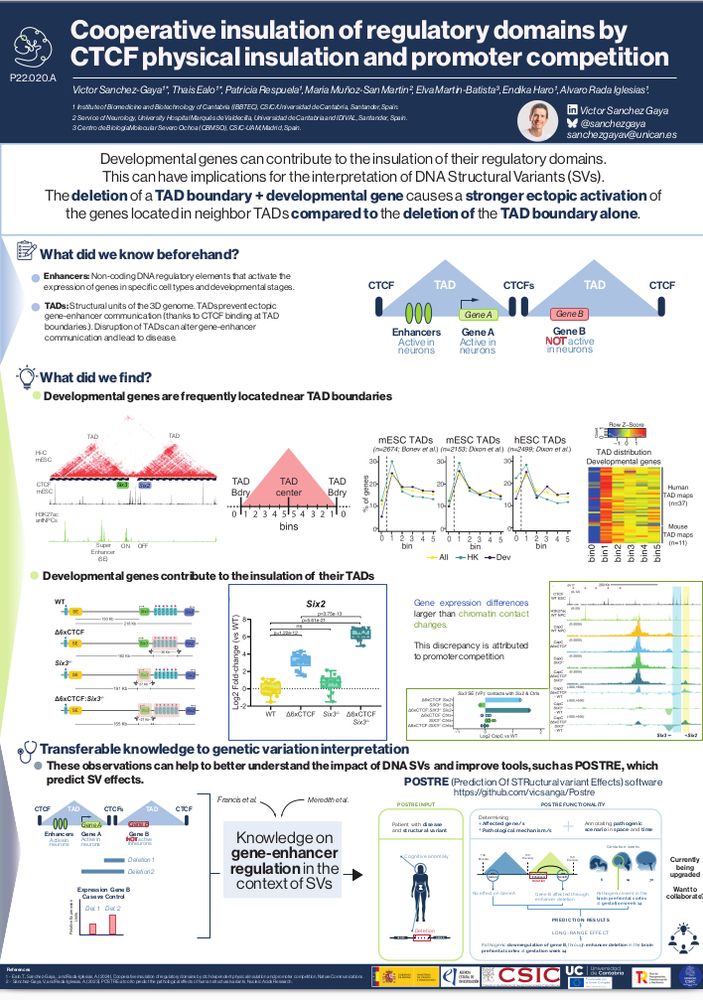

Ready and excited to start with #ESHG2025. If you want to hear about how developmental genes can contribute to the insulation of their own TADs, and the consequences for SV interpretation, find me on P22.020.A poster on Sunday 13:00-14:00. Special thanks to the best host @mattetrovato.bsky.social ❤️
23.05.2025 22:08 — 👍 4 🔁 1 💬 0 📌 2
We are looking to take on an intern at EMBL Rome, for anyone interested in CRISPR and Epigenetics training.
The traineeship is 3-12-months, in Rome (Italy), should start by July, and is supervised by the outstanding @steliostsagkris.bsky.social.
#job #internship; see info below...
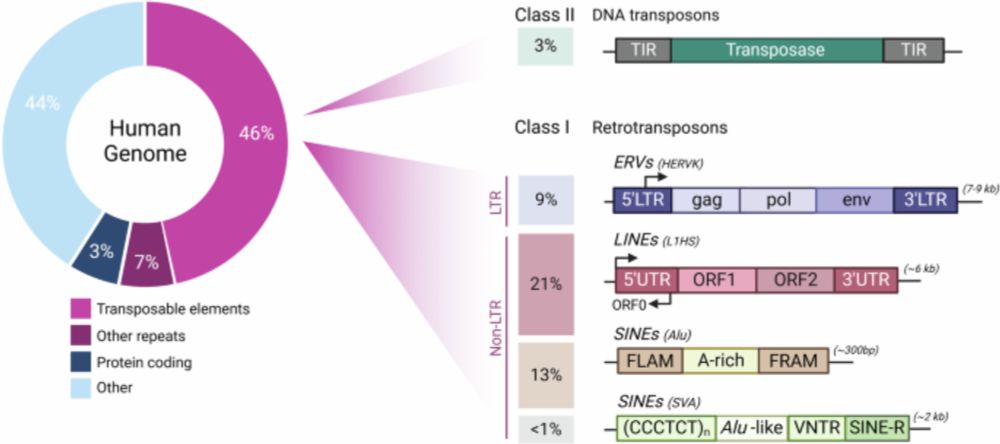
I am excited to share our lab's first review out with in Blood Cancer Journal. We discuss the impact of transposable elements on genome regulation and variation in both normal
haematopoietic processes and haematological cancers. www.nature.com/articles/s41...

Want to work with us on DNA methylation and rare genetic disease? Fully funded PhD project with deadline 16th May:
www.findaphd.com/phds/project...
Excited to collaborate with @hannahlong.bsky.social and Daria Bunina (@uoe-igc.bsky.social/@mdc-berlin.bsky.social).
Please share 🙏
#epigenetics
Many congrats, guys!!! 😎🎉
09.04.2025 09:23 — 👍 0 🔁 0 💬 0 📌 0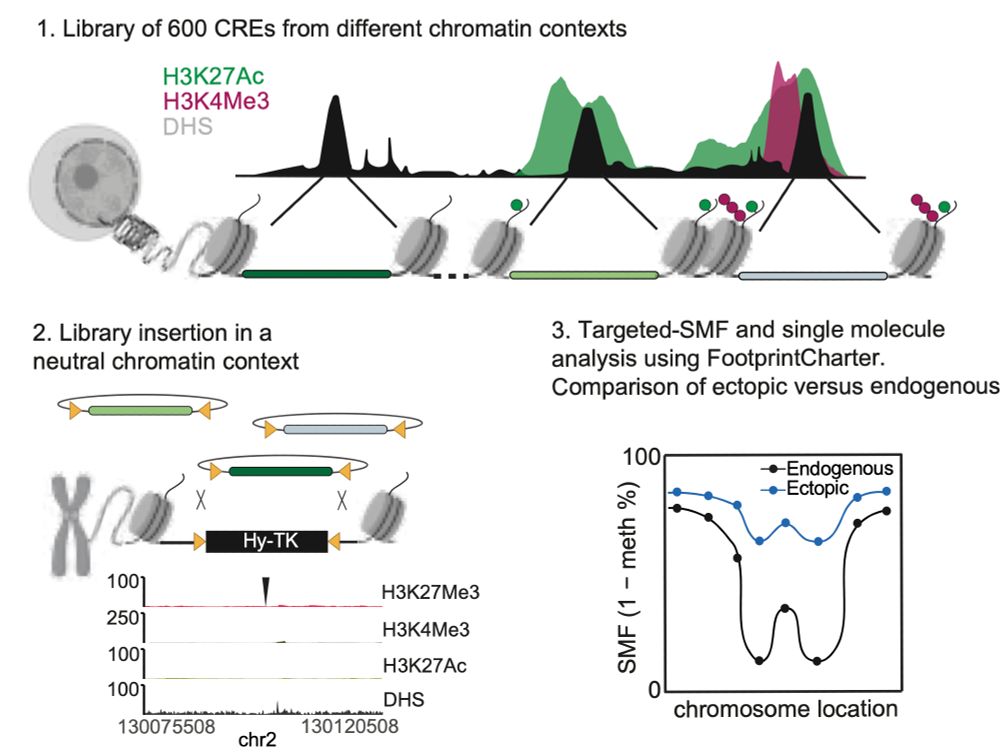
Happy to share the latest story from @arnaudkr.bsky.social's lab @embl.org! With @guidobarzaghi.bsky.social, we used Single Molecule Footprinting to quantify how often chromatin is accessible at enhancers after TF and chromatin environment changes! Check our preprint bit.ly/3XQMFxN + thread ⬇️ 1/11
08.04.2025 13:51 — 👍 78 🔁 33 💬 4 📌 2Super cool to see this out! Many congrats Vale, Guido and everyone involved!! 🥳🥳
09.04.2025 09:21 — 👍 1 🔁 0 💬 0 📌 0
And then, one day before heading to the lab, you stop by uni to hear Shinya Yamanaka sharing the latest on iPSCs research!! Not a bad way to start a Friday...! 😄
#MLS
#MilanLongevitySummit
Check out our latest work on chromatin evolution by @crisnava.bsky.social and @seanamontgomery.bsky.social. Building on our study of histone mark conservation (www.nature.com/articles/s41...), we now explore whether these same marks define similar chromatin states across eukaryotes.
19.03.2025 12:09 — 👍 34 🔁 13 💬 0 📌 0
#CellPlasticity—the ability of cells to change their identity—is vital for tissue growth and repair. But when it goes unchecked, it can fuel #cancer. Our latest study examines how to block #LiverCancer by actively suppressing plasticity. www.nature.com/articles/s41... #CancerBiology
13.02.2025 12:16 — 👍 95 🔁 32 💬 8 📌 4Next Wed 12.02. at 6pm CET I will be speaking about our latest preprint: www.biorxiv.org/content/10.1... and present our findings on how we developed a machine learning model that identified new reprogramming barriers, such as H3K27ac, and our in vivo experiments to test it. See you there!
06.02.2025 12:24 — 👍 18 🔁 10 💬 0 📌 0I am looking for postdocs to work across the fields of #CellSignalling and Chromatin. Background in #Bioinformatics #PTMbiology
#ERC-funded positions - Reach out if you want to discuss.
Please share among potential candidates!
2025 Fragile Nucleosome talks kick off this Wednesday with two great speakers! We'll hear about everything from tardigrades and cryo-EM of nucleosome bound factors to SWI/SNF, LTRs, and ovarian tumors.
You won't want to miss this one, register here! us06web.zoom.us/webinar/regi...
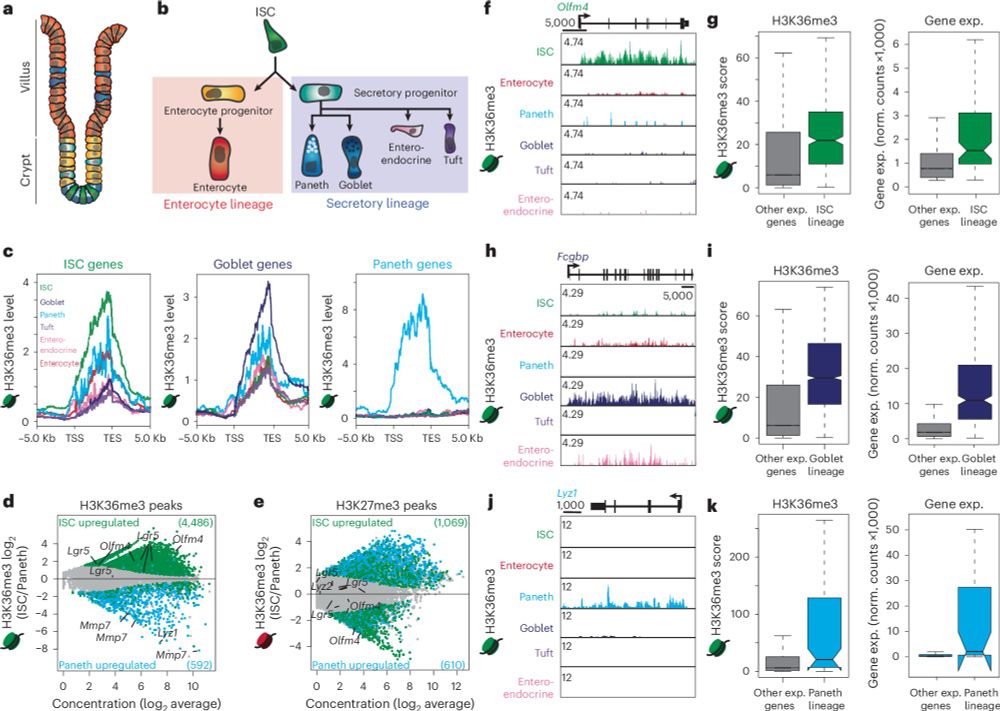
Our new paper defining the role of H3K36 methylation in regulating cell fate and plasticity is available online: rdcu.be/d5AEc
Thanks to our entire team for their tireless work and to the editors at @naturecellbiology.bsky.social for the opportunity!

To kick start the New Year, I'm delighted to share the lab's first preprint - now online:
biorxiv.org/cgi/content/...
See below for a couple of highlights👇
Any feedback is welcome!
"TEsky"
HAPPY NEW YEAR to everyone! 🥂
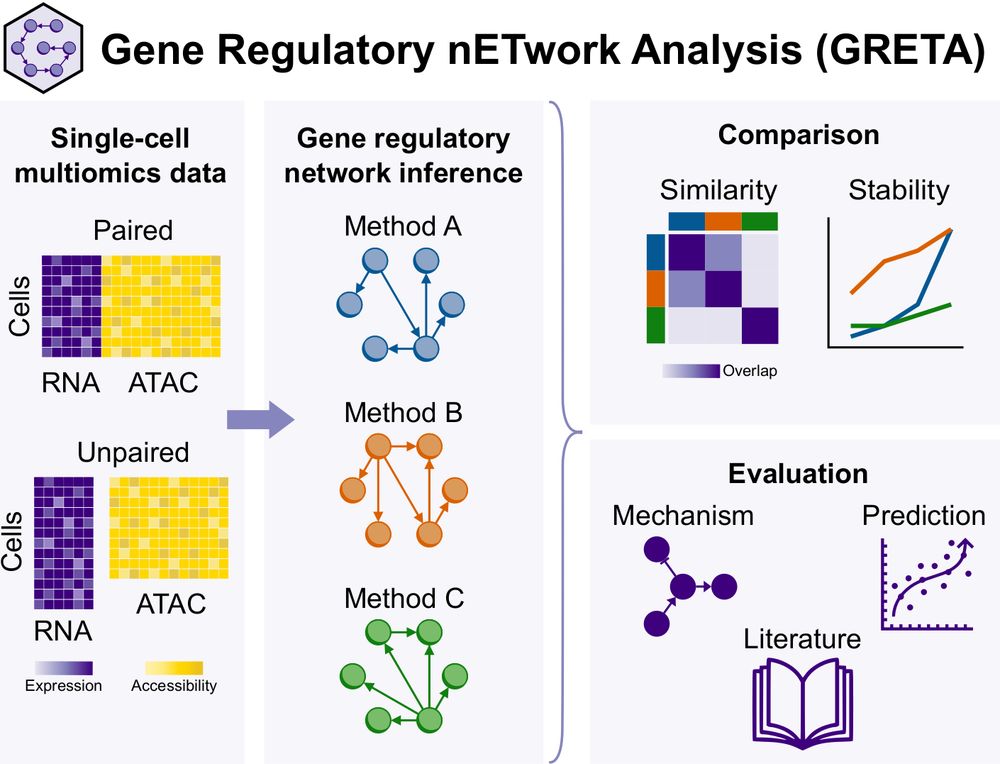
GRETA graphical abstract
We present Gene Regulatory nETwork Analsyis (GRETA), a framework to infer, compare and evaluate gene regulatory networks #GRNs. With it, we have benchmarked multimodal and unimodal GRN inference methods. Check the results here 👇
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta

👋 Hi there ! We are The Lonely Pipette podcast. We are back here to connect with academics. #academicsky
We help scientists do better science by sharing conversations with scientists all around the world, tips to improve the academic ecosystem 🫂
🎧 You can check us here ➡️ bit.ly/TLPtree
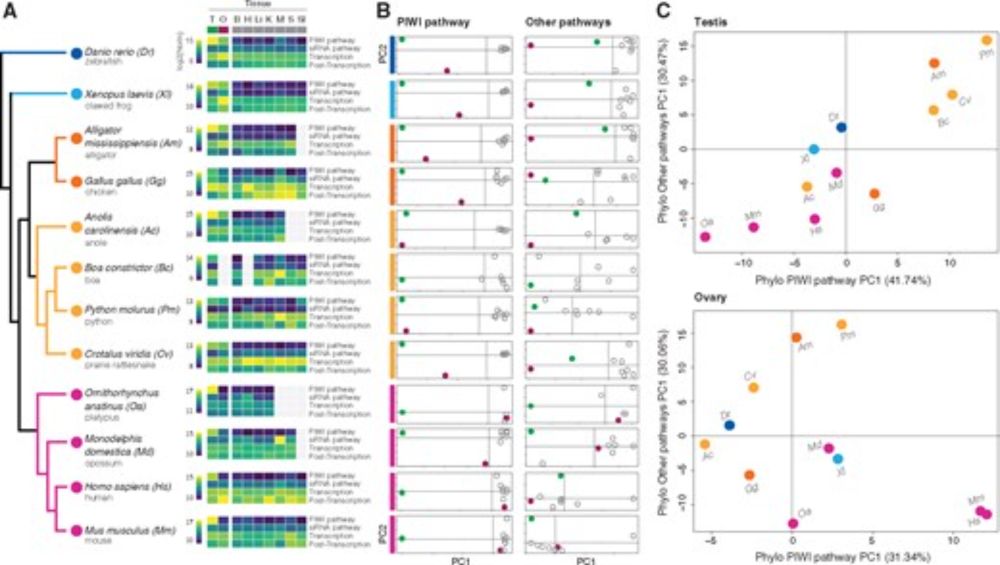
Following up on Nat. Commun. 2018, we analyzed the relationship between TE transcription and expression levels of genes involved in TE silencing in somatic and germline tissues across vertebrates.
doi.org/10.1093/gbe/...
Very nice combinatorics experiment on chromatin IP (ChIP) (one of those like - of course one can do this, why didn't I think of this? - no doubt it is fiddly and complex to get all right) - being able to do ~150 way parallel chip. Gets me thinking about other combinatorics one can do here.
27.11.2024 17:39 — 👍 16 🔁 3 💬 1 📌 0[🔴New episode] What's the ideal recipe to go from 🔲 to ✅?
🎙️ Ana Boskovic, group leader @embl.org shares with us what we might call the “First lab manifesto”
LISTEN NOW 🎧 ► www.buzzsprout.com/1356877/epis...
💌 ► Receive Season 3 in your mailbox : bit.ly/NLTLP

The second preprint presents an experiment workflow and a broadly applicable computional framework to integrate sequencing-based and imaging-based spatial omics data for the SAME tissue slide to construct a single cell spatial omics atlas.
www.biorxiv.org/content/10.1...