Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!
26.10.2025 22:40 — 👍 103 🔁 41 💬 3 📌 5

You asked and we listened... @workshopmlsb.bsky.social is excited to be expanding to Copenhagen, DK at @euripsconf.bsky.social 🎉
Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫
12.09.2025 12:43 — 👍 10 🔁 6 💬 2 📌 2
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 305 🔁 109 💬 14 📌 11

@aithyra.bsky.social Opening Symposium "AI for Life Science" with Nobel Laureate Frances Arnold as keynote speaker in addition to an outstanding line up of speakers on a variety of topics across biological scales and data modalities.
Registration is now open at cemm.at/aithyra-symp...
24.07.2025 19:26 — 👍 19 🔁 9 💬 1 📌 0
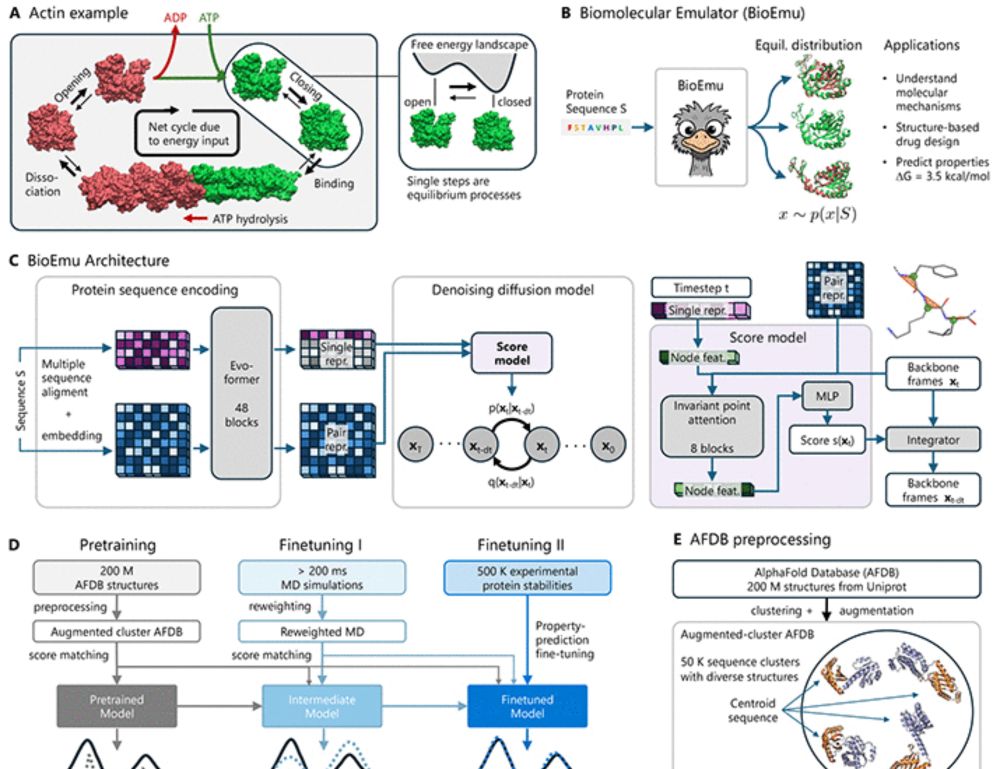
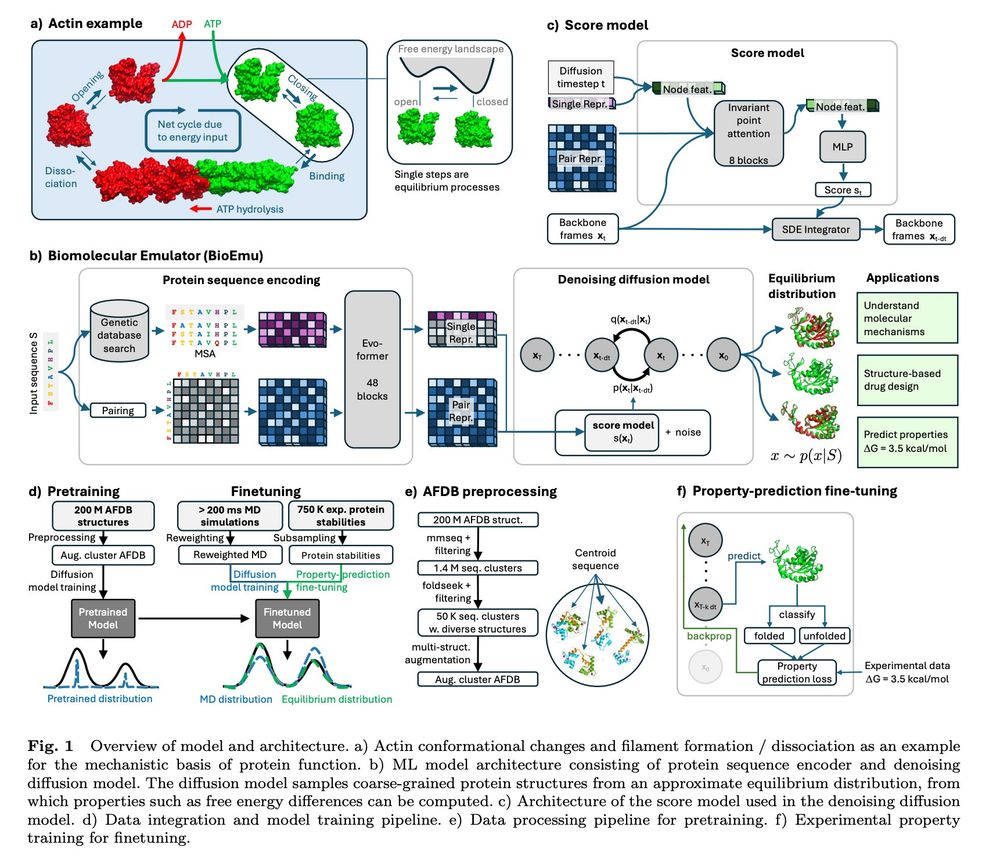
BioEmu now published in @science.org !!
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
10.07.2025 18:57 — 👍 91 🔁 28 💬 6 📌 4
BioEmu is out! Grateful to have had the opportunity to work with such an incredible team on this project 🤗
10.07.2025 20:00 — 👍 5 🔁 2 💬 0 📌 0

This illustration shows the interface of the kinase NEK7 (light blue) and the E3 ubiquitin ligase substrate receptor cereblon (teal). The interaction is facilitated by the small molecule MRT-3486 (orange).
New research in Science greatly expands the potential target scope of molecular glues and should stimulate the development of new small molecules that can selectively target therapeutically relevant proteins for degradation.
Learn more in this week's issue: scim.ag/44uRorm
03.07.2025 18:05 — 👍 54 🔁 17 💬 0 📌 2
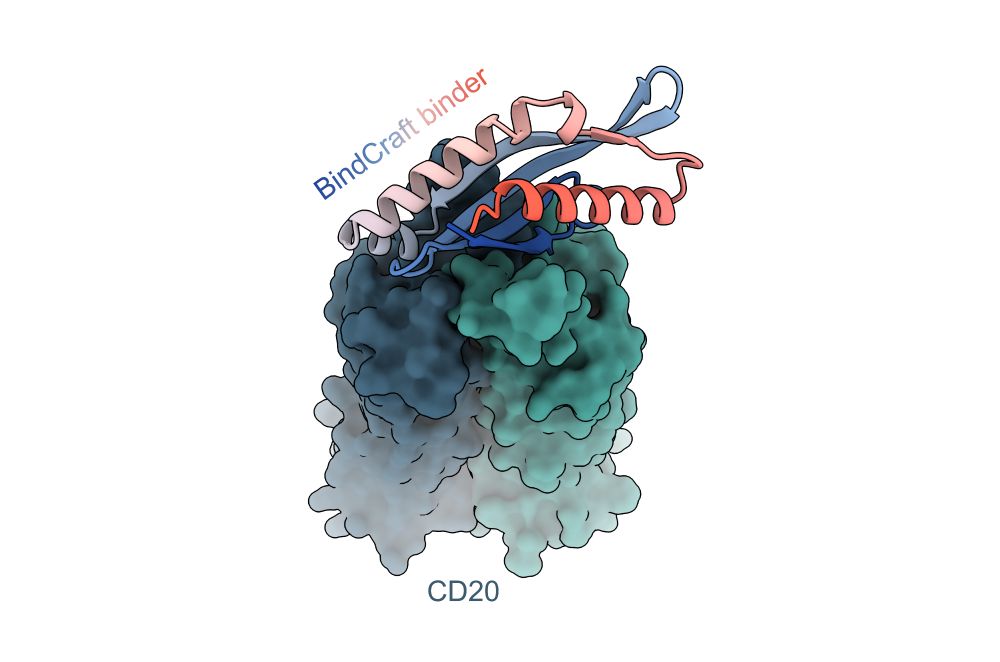
We have written up a tutorial on how to run BindCraft, how to prepare your input PDB, how to select hotspots, and various other tips and tricks to get the most out of binder design!
github.com/martinpacesa...
30.06.2025 19:45 — 👍 138 🔁 55 💬 4 📌 0
Please welcome AITHYRA, the Research Institute for Artificial Intelligence of the Austrian Academy of Science on social media. Follow us and connect via Bluesky and LinkedIn 👋
28.04.2025 07:45 — 👍 17 🔁 5 💬 0 📌 0

Come to see our papers at #ICLR2025 in Singapore
21.04.2025 14:55 — 👍 31 🔁 2 💬 1 📌 0

RAG-ESM logo
📢 Our new preprint is out on bioRxiv! We introduce RAG-ESM, a retrieval-augmented framework that improves pretrained protein language models like ESM2 by making them homology-aware with minimal additional training costs.
🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7
11.04.2025 14:47 — 👍 5 🔁 3 💬 1 📌 1

Google Colab
Run BioEmu in Colab - just click "Runtime → Run all"! Our notebook uses ColabFold to generate MSAs, BioEmu to predict trajectories, and Foldseek to cluster conformations.
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
29.03.2025 09:50 — 👍 102 🔁 42 💬 1 📌 2

Work With Us
The LIT welcomes individuals from all life paths and academic backgrounds. If you are passionate about science we could be the perfect home for you.
I’m looking for a BTA and a Postdoc to help set up the Structural Biochemistry group at the Leibniz Institute for Immunotherapy (LIT) in Regensburg 😀.
We apply synthetic biology and protein design to address key challenges in immunotherapy.
lit.eu/work-with-us/
#SynBio #ProteinDesign #Immunotherapy
13.03.2025 14:50 — 👍 10 🔁 4 💬 0 📌 0
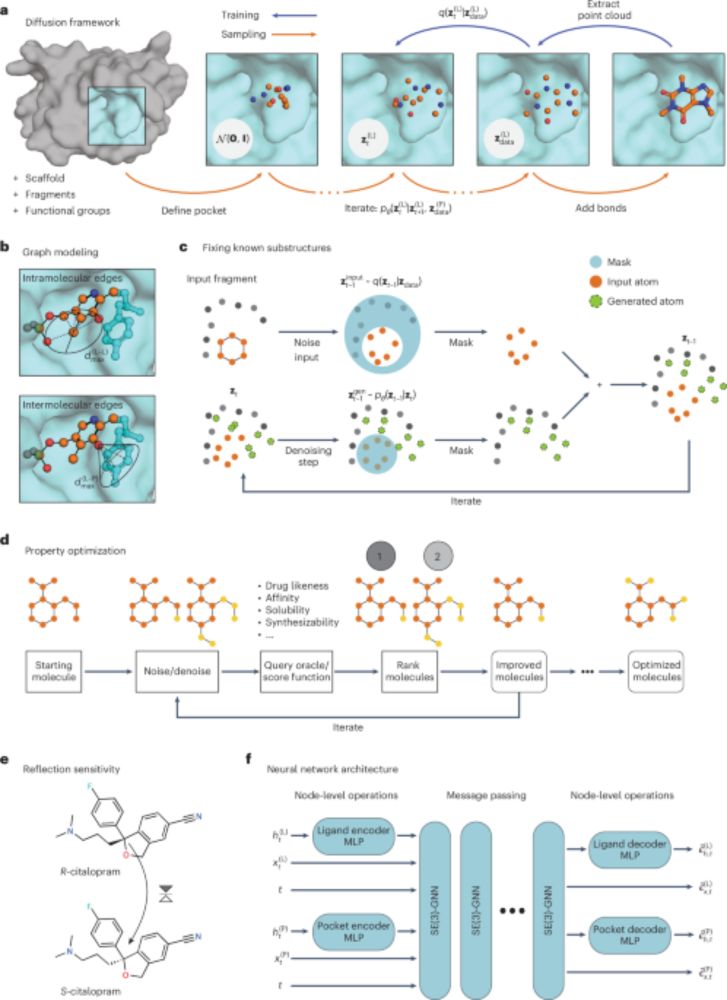
🚨 Check out DrugFlow, our new generative model for structure-based drug design. DrugFlow provides an atom-level confidence score for each designed molecule, and can adjust molecular size on the fly!
Additional details in thread 🧵
#ICLR2025
11.03.2025 15:02 — 👍 23 🔁 7 💬 1 📌 0

Introducing All-atom Diffusion Transformers
— towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team
There are a couple ML ideas which I think are new and exciting in here 👇
10.03.2025 16:19 — 👍 44 🔁 9 💬 1 📌 3

For benchmarking, we placed a lot of emphasis on distribution learning capabilities because this reflects the training objective of generative models. But we also show how downstream preference optimization can be used to further improve molecular properties.
(4/4)
07.03.2025 13:38 — 👍 0 🔁 0 💬 0 📌 0

These models are equipped with a few new features including:
1. protein side chain modeling
2. adaptive ligand sizes
3. confidence score
4. preference alignment
(3/4)
07.03.2025 13:38 — 👍 1 🔁 0 💬 1 📌 0
We (together with @igashov.bsky.social, @adobbelstein.bsky.social, Thomas, @mmbronstein.bsky.social, and Bruno) introduce two new models for target-conditioned drug design in 3D (DrugFlow and FlexFlow), which sample new molecules using a mixed continuous/discrete generative framework.
(2/4)
07.03.2025 13:38 — 👍 3 🔁 1 💬 1 📌 0
The code & camera-ready version of our #ICLR2025 paper on "Multi-domain Distribution Learning for De Novo Drug Design" are now available
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)
07.03.2025 13:38 — 👍 25 🔁 7 💬 2 📌 1
The BioEmu-1 model and inference code are now public under MIT license!!!
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
19.02.2025 20:17 — 👍 103 🔁 39 💬 2 📌 2

We processed the results of the BindCraft user experience poll and we are quite happy with how it turned out. We had over 60 responses from many different users, turns about about a quarter are from industry and a quarter of users run it via Google Colab!
19.01.2025 12:15 — 👍 17 🔁 5 💬 1 📌 0
Our paper on computational design of chemically induced protein interactions is out in @natureportfolio.bsky.social. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia!
t.co/vtYlhi8aQm
15.01.2025 16:37 — 👍 65 🔁 25 💬 1 📌 0
Awesome paper!
BioEmu from MSR: Scalable emulation of protein equilibrium ensembles with generative deep learning www.biorxiv.org/content/10.1...
Looking forward to hearing more explanainers in tomorrow's reading group session on zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
12.01.2025 17:22 — 👍 47 🔁 12 💬 1 📌 0
Super excited to preprint our work on developing a Biomolecular Emulator (BioEmu): Scalable emulation of protein equilibrium ensembles with generative deep learning from @msftresearch.bsky.social ch AI for Science.
www.biorxiv.org/content/10.1...
06.12.2024 08:38 — 👍 441 🔁 147 💬 21 📌 29
@rebeccaneeser.bsky.social @damianosg.bsky.social @pschwllr.bsky.social
21.11.2024 17:35 — 👍 2 🔁 0 💬 0 📌 0
UofT CompSci PhD Student in Alán Aspuru-Guzik's #matterlab and Vector Institute | prev. Apple
MIT biology lab using computational and experimental methods to study protein structure, function, and interactions
https://cen.acs.org/
Sharing the world of #chemistry: News🗞 Research⚗️ Infographics #PeriodicGraphics ℹ Pictures #CENChemPics 📷 Comics #SketchChemistry 🗯 #FluorescenceFriday ❇️
machine learning researcher @ Apple machine learning research
the Open Molecular Software Foundation is dedicated to the creation and implementation of the latest and greatest open comp.chem software.
Building generative models for high-dimensional science and engineering.
Assistant prof. @CarnegieMellon & affiliated faculty @mldcmu, previously instructor @NYU_Courant, PhD jointly @Harvard and @MIT
https://nmboffi.github.io
junior fellow at @Harvard.edu, incoming prof at @HSEAS and @Kempnerinstitute.bsky.social studying machine learning and its applications to nature and the sciences
Professor at EPFL. Une mathémaphysinformaticienne. Passionate mushroom hunter. Tamer of two little dragons.
PhD @EPFL with Andrea Ablasser & Didier Trono
Focusing on Innate Immunity, Epigenetics and Gene Regulation, trying to not be distracted by the rest of biology
We are a structural biology lab located both at EPFL in Lausanne and at the Friedrich Miescher Institute in Basel.
thomalab.org
www.epfl.ch/labs/upthomae
Postdoc @labthoma.bsky.social at EPFL, ISREC
Former PhD @guillaumediss.bsky.social at @FMIscience.bsky.social
From Mutations 🧬 to Meanings 🙌🏻
Lab account for AG Niopek at the University of Heidelberg IPMB
Protein Engineering | Allostery | CRISPR | Optogenetics | ML
Account is managed by PhD Students
https://Niopeklab.de/
Machine Learning Prof @UCSanDiego, #Physics-Guided #AI, MIT TR-35 Innovator. Website: roseyu.com
Scientist with a passion for Synbio | Protein engineering | Optogenetics | ML
www.niopeklab.de/mathony-lab/
AI4science research, density functional theory @ Microsoft Research Amsterdam. PhD on generative modeling, flows, diffusion @ Mila Montreal
PhD student at AstraZeneca and Chalmers University working on generative models for drug design
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Assistant professor in Data Science and AI at Chalmers University of Technology | PI: AI lab for Molecular Engineering (AIME) | ailab.bio | rociomer.github.io