With this, the last bit of my PhD at @embl.org is finally out!
We developed salad (sparse all-atom denoising), a family of blazing fast protein structure diffusion models.
Paper: nature.com/articles/s42256-…
Code: github.com/mjendrusch/salad
Data: zenodo.org/records/14711580
1/🧵
24.09.2025 11:58 — 👍 24 🔁 9 💬 1 📌 0
🎉🎉 congrats Michael!!
24.09.2025 16:35 — 👍 1 🔁 0 💬 1 📌 0
Happy to hear that, thank you!
22.08.2025 06:55 — 👍 0 🔁 0 💬 0 📌 0
Thanks Julian!
05.08.2025 08:17 — 👍 1 🔁 0 💬 0 📌 0
Work by @bene837.bsky.social @jmathony.bsky.social @dominikniopek.bsky.social @uniheidelberg.bsky.social
04.08.2025 15:26 — 👍 12 🔁 5 💬 0 📌 0

Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Nature Methods - ProDomino is a machine leaning-based method, trained on a semisynthetic domain insertion dataset, to guide the engineering of protein domain recombination.
Publication alert: Our paper on domain insertion predictions in proteins is now out in @natmethods.nature.com in its final form: rdcu.be/ey7w3
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...
04.08.2025 12:28 — 👍 8 🔁 4 💬 1 📌 1
I'm very excited to finally share the main work of my PhD!
We explored the evolutionary dynamics of gene regulation and expression during gonad development in primates. We cover among others: X chromosome dynamics (incl. in a developing XXY testis), gene regulatory networks and cell type evolution.
20.06.2025 08:35 — 👍 14 🔁 5 💬 1 📌 0
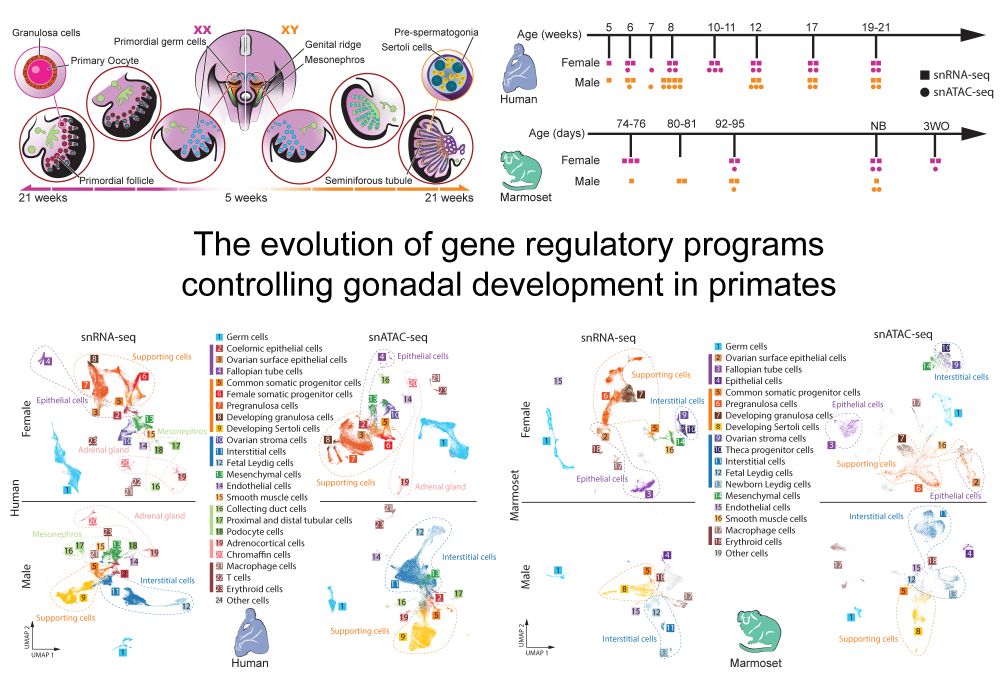
We are delighted to share our new preprint “The evolution of gene regulatory programs controlling gonadal development in primates” www.biorxiv.org/content/10.1...
20.06.2025 08:12 — 👍 90 🔁 36 💬 4 📌 1
Congrats Michael, great work as always!
20.06.2025 06:10 — 👍 1 🔁 0 💬 1 📌 0
Happy to announce the first paper from my PhD at Korbel group at @embl.org has finally been published:
embopress.org/doi/full/10.1038…
Collaborating with @typaslab.bsky.social, @hennig-lab.bsky.social and the EMBL PEPCF, we designed de novo inhibitors to a bacterial phage defense system 1/🧵
19.06.2025 08:10 — 👍 30 🔁 7 💬 2 📌 3

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
Inspired by how nature evolves trigger responsiveness through alternating pressures, we are excited to present POGO-PANCE and RAMPhaGE:
Phage-assisted evolution platforms for engineering allosteric protein switches under dynamic selection.
Preprint: doi.org/10.1101/2025...
13.06.2025 13:29 — 👍 14 🔁 3 💬 1 📌 2
Find a longer thread about our work here:
bsky.app/profile/niop...
13.06.2025 14:12 — 👍 1 🔁 0 💬 0 📌 0
New Preprint on phage-assisted evolution and retron-mediated mutagenesis for protein optimization.
Many congrats to @neuroscinikolai.bsky.social for this heroic effort and all other lab members involved!
13.06.2025 11:03 — 👍 7 🔁 1 💬 1 📌 0
Huge congrats to all authors, especially to co-first authors @ann-sophiekroell.bsky.social and Kira Hoffmann, who did an outstanding job!
03.05.2025 11:03 — 👍 1 🔁 0 💬 0 📌 0
Finally, we also show efficient thermal protein control using another (chemo-)receptor domain as insert. Our work expands thermogenetics to the allostreric control of proteins.
03.05.2025 11:03 — 👍 0 🔁 0 💬 1 📌 0
The insertion of our LOV2 variants into transcription factors, Cas9 and an RNA-binding protein enables the thermogenetic control of various cellular processes. We expect the approach to be easily tranferrable to the various AsLOV2-based tools out there.
03.05.2025 11:03 — 👍 0 🔁 0 💬 1 📌 0
New paper alert! We introduce the modular allosteric thermo-control of protein activity. Employing the AsLOV2 domain and mutants thereof as thermoreceptors, we engineered diverse hybrid proteins, whose activity can be controlled by small temperature changes (37-40/41 °C).
doi.org/10.1101/2025...
03.05.2025 11:03 — 👍 13 🔁 5 💬 3 📌 0
It was a pleasure to meet all of you and discuss science! Many thanks again for the invitation.
22.04.2025 15:17 — 👍 1 🔁 0 💬 0 📌 0

PyFuRNAce: An integrated design engine for RNA origami
To realize the full potential of RNA nanotechnology and RNA origami, user-friendly design tools are needed. Here, we present pyFuRNAce, an open-source, Python-based software package with a graphical u...
Hey #RNA world, we are excited to share our RNA design engine #pyFuRNAce - an integrated tool for RNA origami experts and novices alike. Kudos to @monari-luca.bsky.social @floppleton.bsky.social et al! Preprint: www.biorxiv.org/content/10.1... Try it out at pyfurnace.de Feedback welcome!
20.04.2025 12:29 — 👍 50 🔁 14 💬 0 📌 1

We have an exciting new PhD opportunity!
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.
01.04.2025 11:52 — 👍 13 🔁 11 💬 0 📌 1


What a week! I defended my PhD on Monday, and now my first first-author paper was published in @science.org.
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛
14.02.2025 18:27 — 👍 26 🔁 2 💬 2 📌 1
Congrats Michael! Impressive work!
07.02.2025 07:44 — 👍 1 🔁 0 💬 1 📌 0
Thanks Nick! It was a lot of fun to work with you and all others :)
16.12.2024 17:32 — 👍 1 🔁 0 💬 0 📌 0
I am super excited and grateful to be funded by the BW-Stiftung within the Postdoc Elite Program. Looking forward to bringing exciting new research from the drawing board into the lab. Many thanks to all my amazing colleagues for all the continued collaboration and support!
09.12.2024 12:20 — 👍 5 🔁 0 💬 0 📌 0
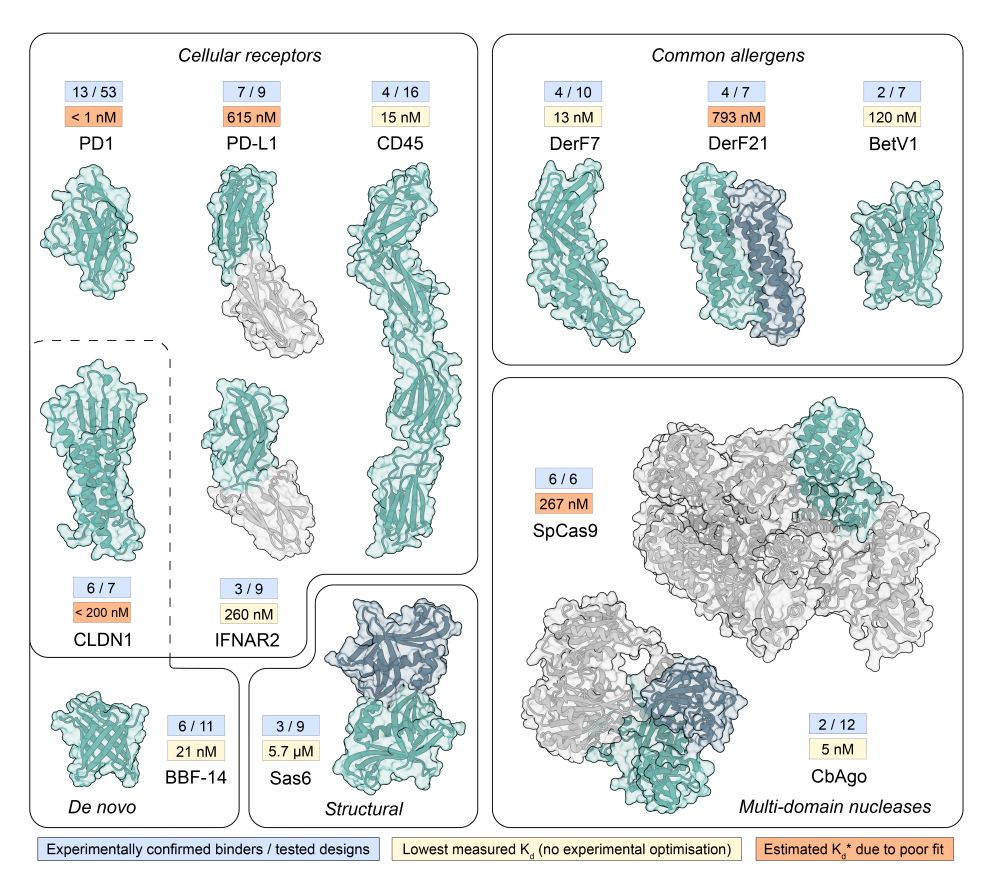
We updated our BindCraft preprint with lots of new exciting results! We release all our binder sequences and models, include more in silico analysis, novel design targets, and present AAV retargeting to specific cell types using de novo binders!
www.biorxiv.org/content/10.1...
08.12.2024 08:26 — 👍 128 🔁 31 💬 1 📌 3
Communicating in the Molecular Language of Life.
DFG funded Cluster of Excellence at the University of Freiburg, Germany
#LifeSciences #Research #Freiburg
Website: https://www.cibss.uni-freiburg.de/
Legal Notice: https://www.cibss.uni-freiburg.de/imprint
Unraveling plant growth at a subcellular level @biologyunifreiburg.bsky.social @dompsfr.bsky.social @uni-freiburg.de
Speaker @cibss.bsky.social
Senior editor @elife.bsky.social
www.plantscience.de
PI at the University of Freiburg, interested in molecular microbiology and all things related to #archaea
Bio-Acousto-Magneto-Neuro-Chemical Engineer
Max Delbrück Professor @Caltech
Investigator @HHMI
Fulbright-Tocqueville Chair @PhysMedParis @ESPCI_Paris '24-5
shapirolab.caltech.edu
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
PhD student @UniHeidelberg | Kaessmann lab | UTokyo alumn
PhD student @EPFL 🇨🇭
ML & computational biology 🤖🧬⚛️
Group leader @UniHeidelberg, @IPMB;
Chemical Biologist; RNA imaging 🦠 Super-resolution 🔬Fluorescent probes 🌈Alumni |@UChicago
Group Website: www.sunbulgroup.com
The ZMBH - the Center for Molecular Biology of Heidelberg University - is a center for research and higher education in molecular biology and biomedicine. Our research aims at fundamental questions of molecular biology.
PhD Student @ Schwille Lab, MPIB. Interested in the synthesis of protein design and synthetic cells. Eventually aiming to create life from scratch.
Assistant Professor at @UCSF_BTS | @HHMINEWS HGF | | Loving membrane proteins every day https://www.wcoyotelab.com
Cell cycle, mitosis, nuclear pores, ubiquitin, disease.
Group leader @igbmc.bsky.social, research director DR1 @cnrs.fr, Strasbourg, France.
Alumna @JagiellonskiUni.bsky.social @impvienna.bsky.social @ethz.ch
Hobby cook and occasional runner.
PhD student at Bugaj Lab UPenn | synbio | protein engineering | optogenetics | thermogenetics | systems bio | love weird dynamics, information flow, networks | BcLOV4 | looking for postdoc positions
RNA and DNA origamist
Currently searching for postdoc positions
PhD in @kgoepfrich.bsky.social working on building a syncell from RNA/DNA nanotech
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Cell biologist, Assistant professor @ Erasmus MC, Optical Imaging Centre (OIC). Postdoc alumnus @ UCSF. Interested in cytoskeleton dynamics, neuronal growth cones, live cell microscopy, optogenetics. Opinions are my own.
Chemical Biologist with passion for Cancer Research, Targeted Protein Degradation & Transcription.Scientific Founder of Proxygen & Solgate. Pats Fan. Dad.
winter-lab.com












