16/ Thank you to my co-first authors @phil-fradkin.bsky.social @heyitsmeianshi.bsky.social, all the authors who got these datasets together (especially Divya Koyyalagunta), and my mentor @quaidmorris.bsky.social / Phil’s mentor @bowang87.bsky.social! Also special thanks to the MSK HPC / NSF-GRFP 💪🏽
15.07.2025 19:08 — 👍 0 🔁 0 💬 0 📌 0
15/ The great news going forward is that mRNABench is 🚀open source, 🪶lightweight, and 🤖modular!
We made it easy to:
📊 Add datasets
🖥️ Add new models
📈Benchmark reproducibly
We hope you will give it a try, and if you have any comments or ideas, feel free to reach out!
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
14/ Conclusions
1. genomic sequences != language -> we need training objectives suited to the heterogeneity inherent to genomic data
2. relevant benchmarking tasks and evaluating generalizability is important as these models start to be used for therapeutic design
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
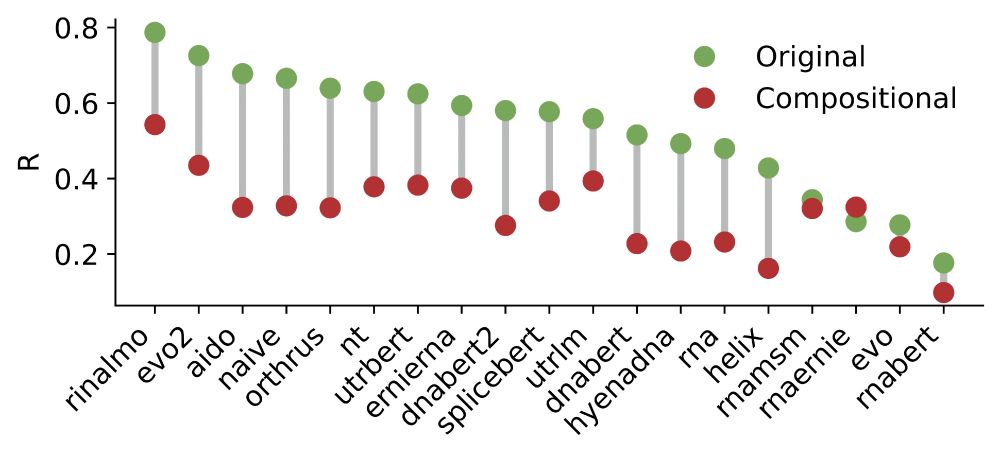
13/ ❌ All models failed badly at this test, with significant drops in Pearson correlation when compared to a random data split, indicating that they don’t fully understand how these regulatory features interact.
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
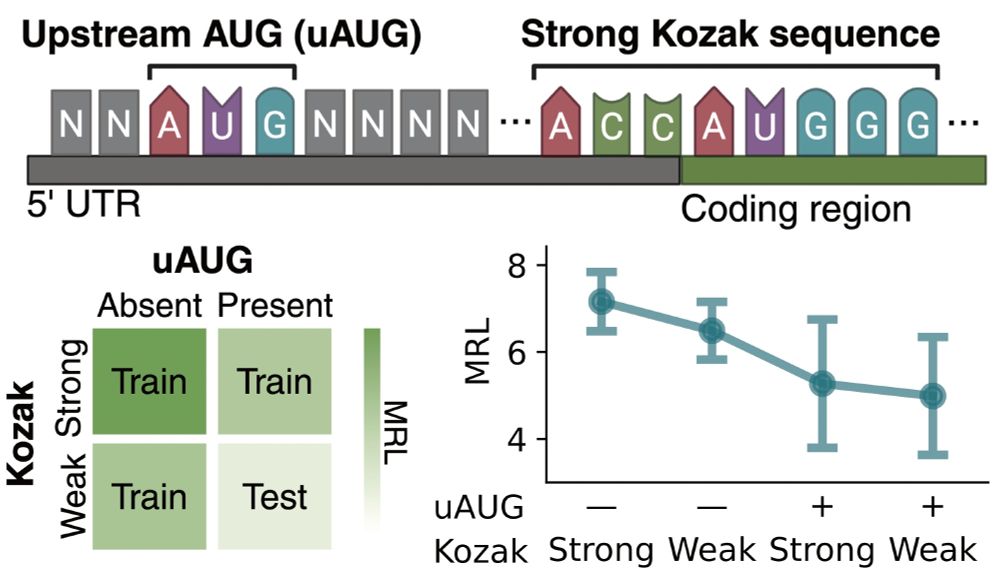
12/ For this task, we used a dataset that measures mean ribosome load when the 5’ UTR sequence is varied.
We know that uUAGs reduce translation, and strong Kozak sequences enhance it, so we trained linear probes using 3 subsets of these features and tested on the held out set.
15.07.2025 19:08 — 👍 2 🔁 0 💬 1 📌 0
11/ Finally, we wanted to evaluate whether current SSL models are compositional ➡️ do they understand how sequence elements that they have seen before interact when combined?
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
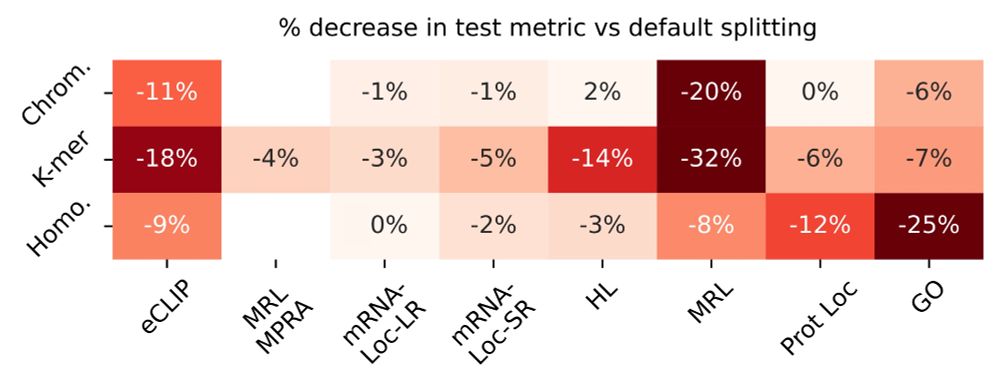
10/ We also show that random data splits (vs biologically-aware data splits) inflate model performance because structurally or functionally related sequences end up in both training and test sets, overestimating model generalization. This drop is highly task dependent.
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
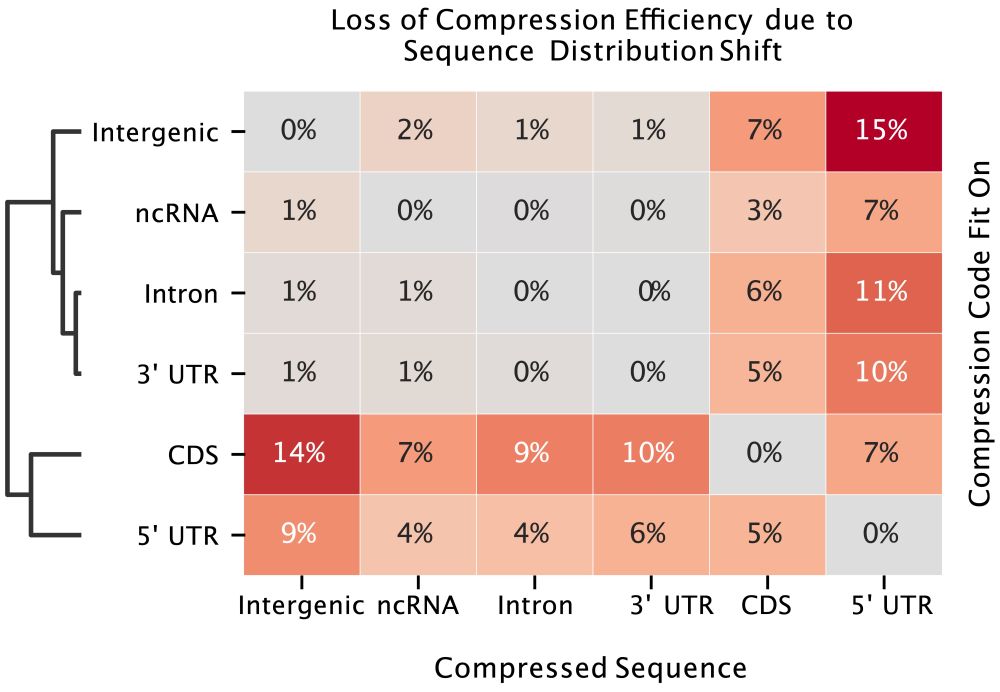
9/ From a compression standpoint, sequence composition differs significantly between genomic regions, explaining why ncRNA models and models trained largely on intronic or intergenic regions might poorly on mRNA specific tasks
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
8/ We trained a simple data compressor (using Huffman encoding) to measure how “similar” different parts of the genome are, by training the compressor on each region (CDS, 5’ UTR, 3’ UTR, introns, etc) and then trying to compress the other regions.
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
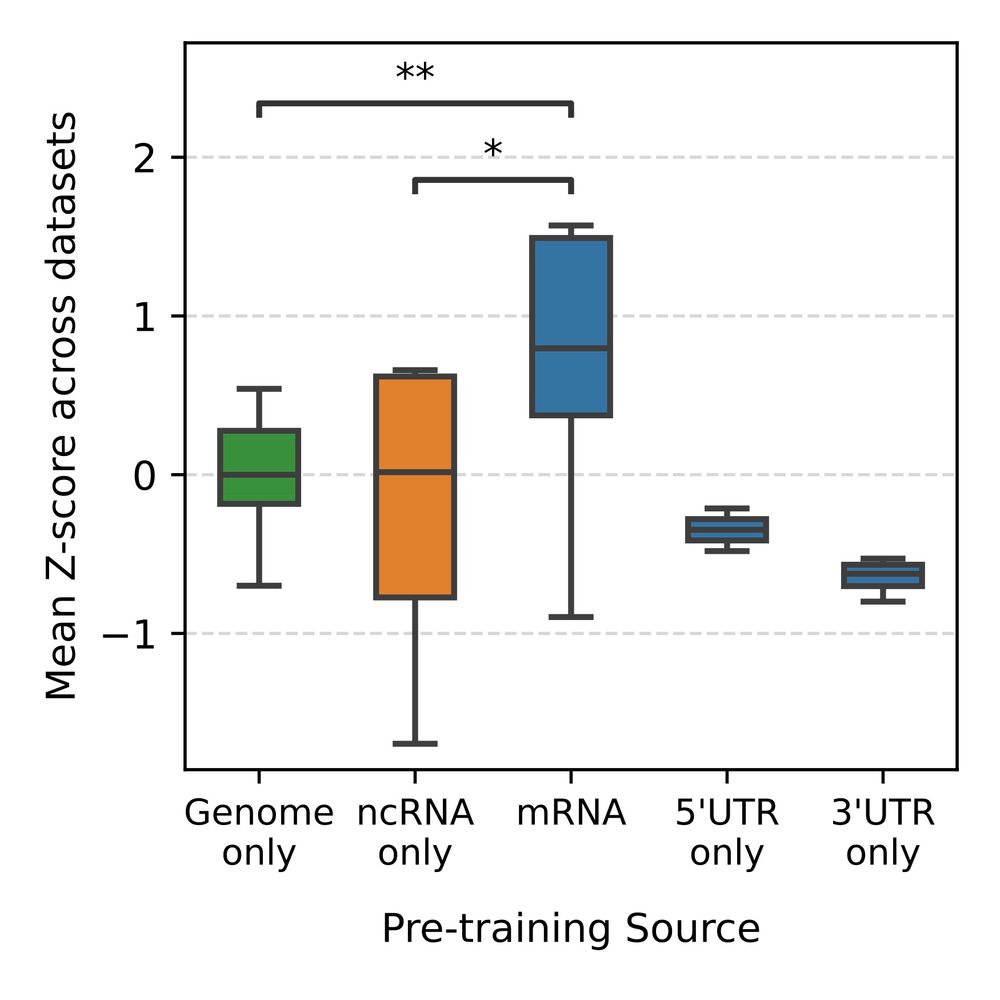
7/ Taking a step back, we also wanted to explore why models pretrained on DNA or ncRNA perform so poorly on mRNA specific tasks
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
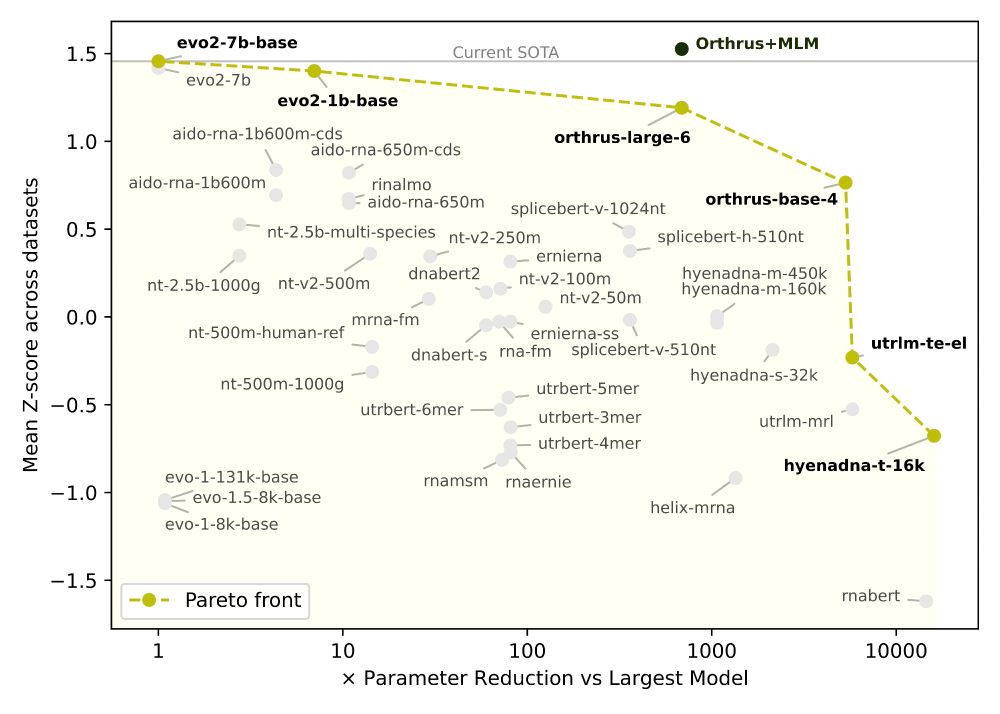
6/
✅ Orthrus+MLM matches/beats SOTA on 6/10 datasets without increasing the training data or significantly increasing model parameters
📈 Pareto-dominates all models larger than 10 million parameters, including Evo2
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
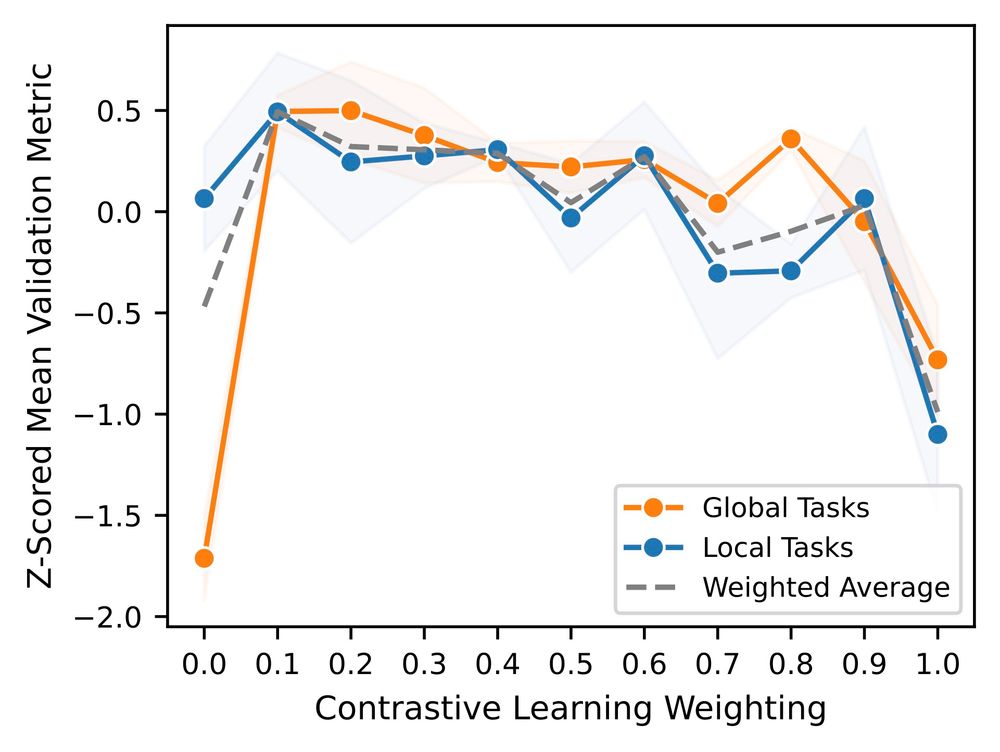
5/ To address this, we pretrained a joint contrastive + MLM Orthrus variant, and investigated the optimal ratio between these two objectives.
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
4/ Orthrus, unlike the other models, uses a contrastive learning objective (which has been shown to yield worse performance on finer resolution tasks). In line with this, we notice that Orthrus underperforms on nucleotide level (local) tasks vs transcript-wide (global) tasks.
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
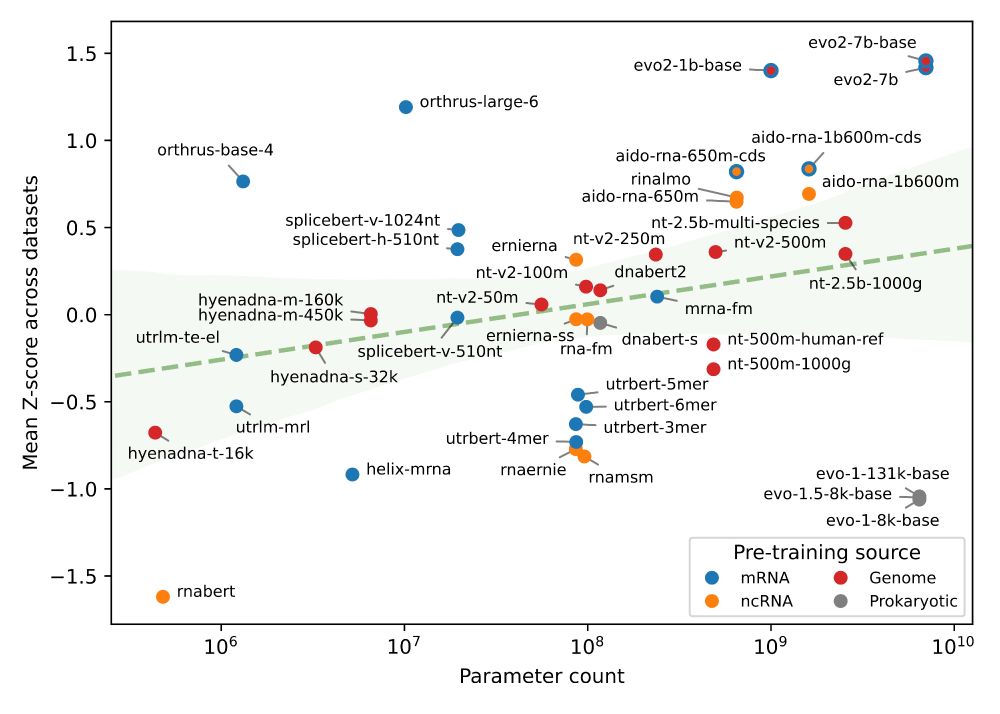
3/ As expected, we see that scaling parameter count generally leads to performance improvements, with Evo2 7 billion performing the best overall
Interestingly, we noticed that the 2nd best model, Orthrus, was not far off compared to Evo2 despite having only 10 million parameters
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
2/ We benchmarked nearly all public models trained on DNA (e.g. HyenaDNA), ncRNA (e.g. AIDO.RNA), mRNA (e.g. Orthrus), and across all of the above (Evo2), evaluating them via linear probes.
In total, we conducted over 135,000 total experiments!
So what did we learn? 👇🏽
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
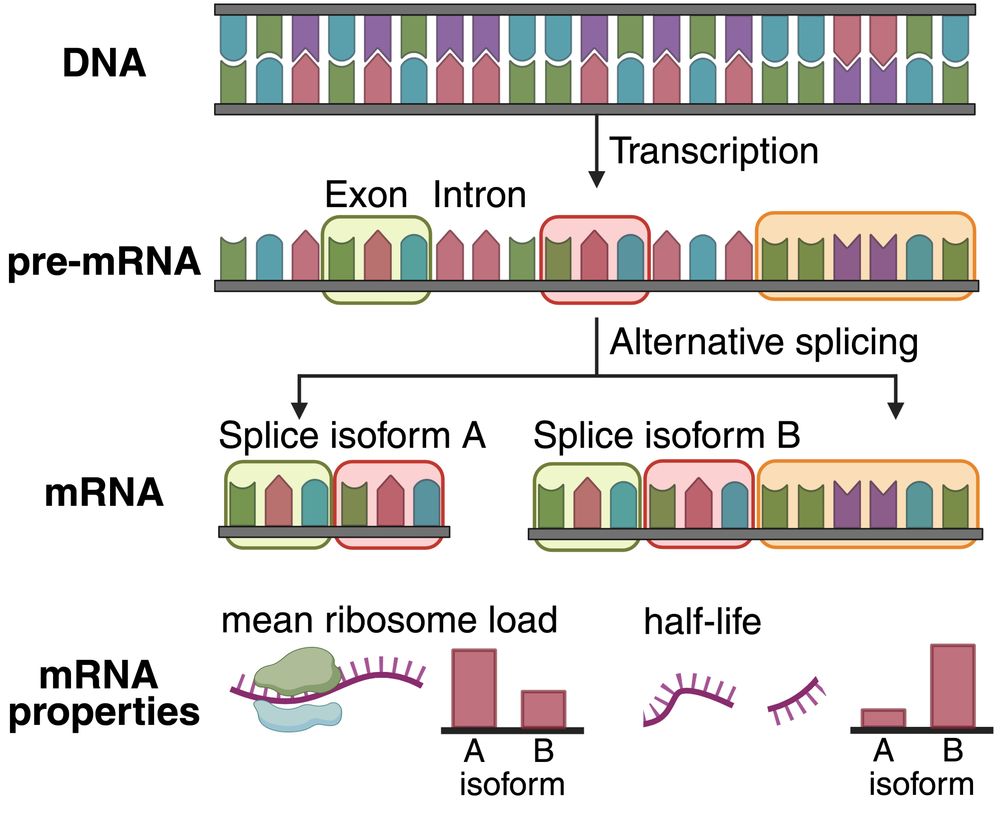
1/ Existing DNA/RNA benchmarks focus on tasks that are either not predictable from mRNA sequence or are structure based. In mRNABench, we brought together 10 datasets that focus on salient mRNA properties and function, like mean ribosome load, localization, half life, etc
15.07.2025 19:08 — 👍 0 🔁 0 💬 1 📌 0
Computational biology, cancer genomics, gene regulation and machine learning.
Postdoc @ MSKCC, previously @Duke
I use robots and biophysics to study cancer therapies in the Chodera Lab at Memorial Sloan Kettering Cancer Center. Views are my own.
Applied ML in living things.
PhD-ing CompBio @MSK w Wesley Tansey // jhu cs/bme
Comp Bio PhD Student in Kushal Dey and Thomas Norman labs at MSK Cancer Center. Interested in gene regulation and sports
PhD student at MSKCC Lareau Lab interested in computational protein design for cancer therapies
PhD student in Comp Bio studying the adaptive immune system
Tri-I CBM PhD Student interested in single-cell modeling
Researcher with the Ashley Laughney and Caleb Lareau
NSF GRFP Fellow
computational biology phd student @weill cornell & msk
CompBio PhD student in Sohrab Shah and Jian Carrot-Zhang labs @ MSK
The Office of Scientific Education & Training at Memorial Sloan Kettering serves as an academic resource for current and prospective scientific and clinical trainees.
Postdoc in Computational Biology @Buenrostro Lab | 3D Genome & Epigenomics | PhD @Leslie Lab
PhD Student in Comp Bio @MSK in Leslie and Dey labs. DNA sequence to function models
PhD Student @ University of Toronto
Building foundation models for genomics!
Born too late to explore the Earth
Too early to explore the Galaxy
Just in time to model the cell and climb some rocks
PhD @ UofT and Vector Institute
Postdoctoral researcher at Blencowe’s lab, University of Toronto 🇨🇦. PhD, University of Cambridge - Wellcome Sanger Institute 🇬🇧. Alternative splicing and Bioinformatics 🧬💻🦾
https://scholar.google.cl/citations?user=PTLeXysAAAAJ&hl=en
Computational biology, machine learning, AI, RNA, cancer genomics. My views are my own. https://www.morrislab.ai
He/him/his
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Professor, Programmer in NYC.
Cornell, Hugging Face 🤗
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app










