
Interesting new @medrxivpreprint.bsky.social study, bearing on the interpretation of GWAS results:
“Common and rare genetic variants show network convergence for a majority of human traits” 🧪🧬
www.medrxiv.org/content/10.1...
@geparada.bsky.social
Postdoctoral researcher at Blencowe’s lab, University of Toronto 🇨🇦. PhD, University of Cambridge - Wellcome Sanger Institute 🇬🇧. Alternative splicing and Bioinformatics 🧬💻🦾 https://scholar.google.cl/citations?user=PTLeXysAAAAJ&hl=en

Interesting new @medrxivpreprint.bsky.social study, bearing on the interpretation of GWAS results:
“Common and rare genetic variants show network convergence for a majority of human traits” 🧪🧬
www.medrxiv.org/content/10.1...

Delighted to share our latest work deciphering the landscape of chromatin accessibility and modeling the DNA sequence syntax rules underlying gene regulation during human fetal development! www.biorxiv.org/content/10.1... Read on for more: 🧵 1/16 #GeneReg 🧬🖥️
03.05.2025 18:27 — 👍 126 🔁 59 💬 2 📌 3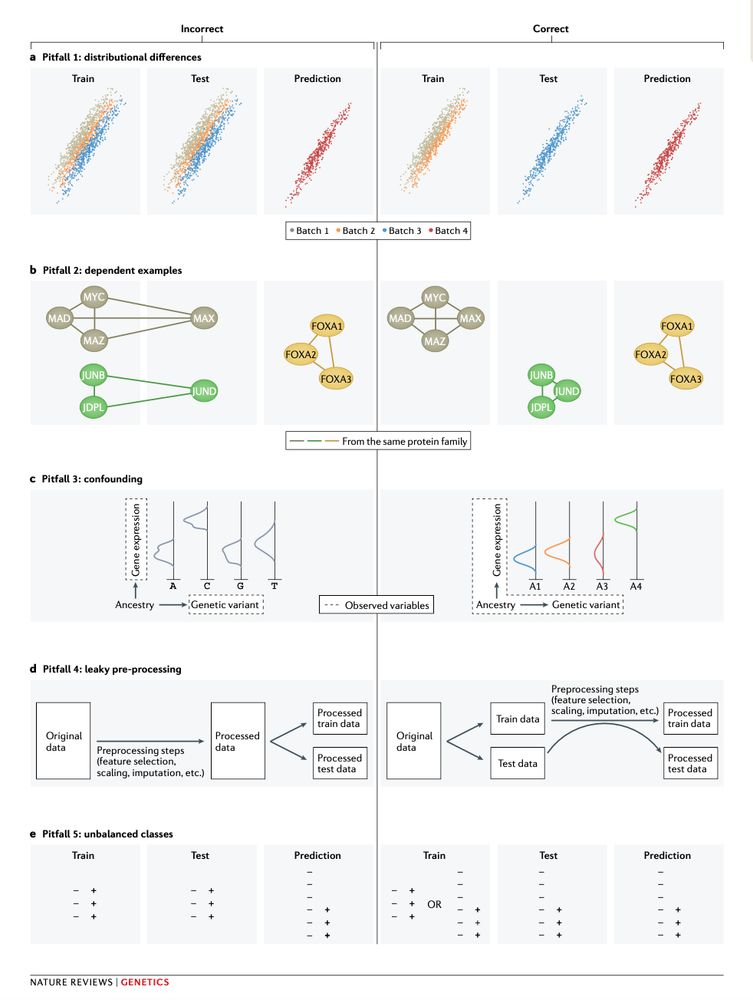
Machine learning (ML) is revolutionizing genomics, but common pitfalls can lead to misleading results. Here's a thread on how to avoid them 🧵
24.01.2025 14:45 — 👍 20 🔁 4 💬 1 📌 1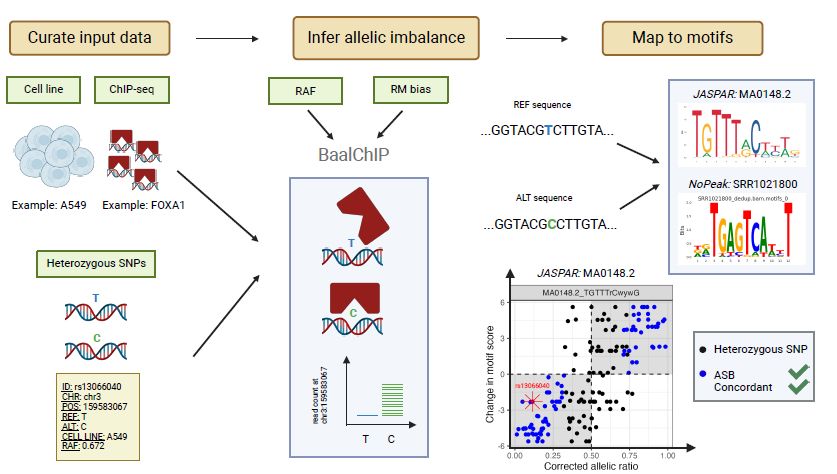
Description of image: Workflow for identifying high-quality ASBs through baal-nf. ChIP-sequencing data from genotyped cell lines is used to infer ASBs at heterozygous SNPs using BaalChIP. Identified ASBs are further characterized by mapping them to JASPAR and NoPeak motifs for a given TF. High-quality ASBs show concordance in the corrected allelic ratio and motif score difference.
What DNA variants alter transcription factor (TF) binding affinity? Here, we describe a new workflow baal-nf. We used it to trawl through 6017 ChIP-Seq data sets for 558 TFs and 46 genotyped cell lines, finding 298,783 allele-specific binding sites (ASBs).
www.biorxiv.org/content/10.1...
1/2
Very excited to announce that the single cell/nuc. RNA/ATAC/multi-ome resource from ENCODE4 is now officially public. This includes raw data, processed data, annotations and pseudobulk products. Covers many human & mouse tissues. 1/
www.encodeproject.org/single-cell/...
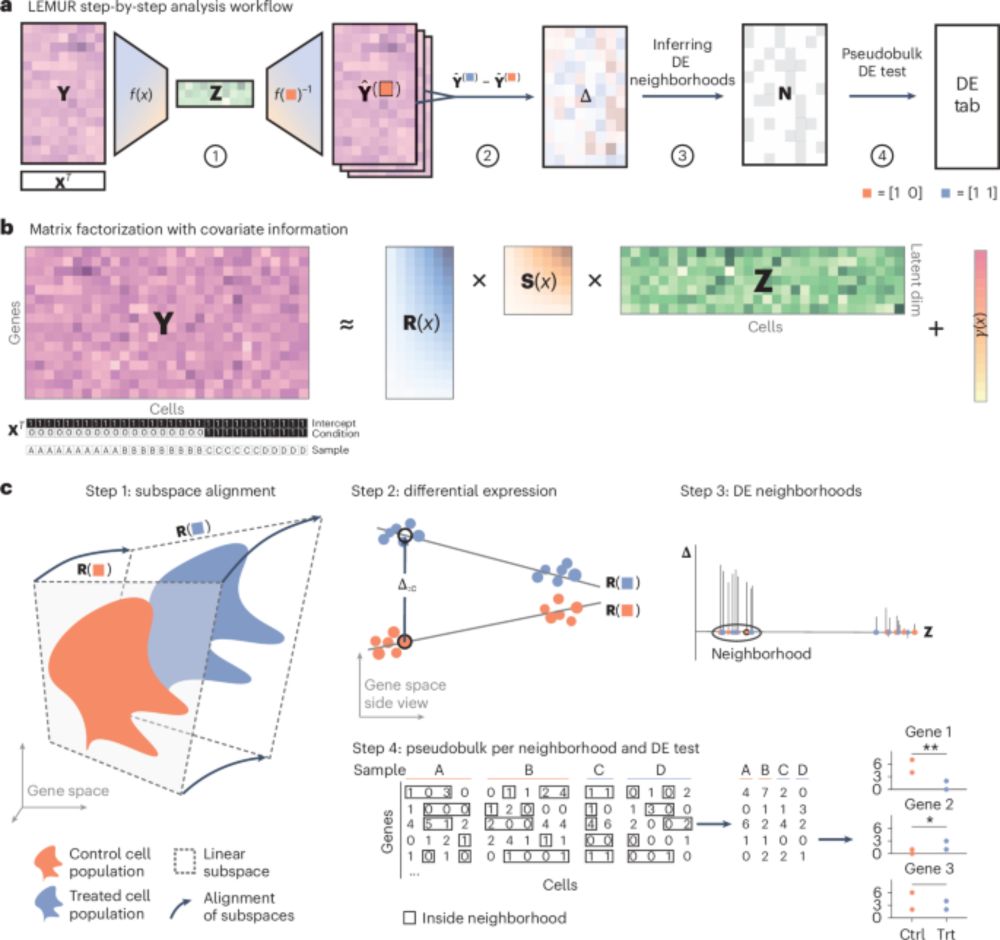
How to do differential expression with scRNAseq data? State of the art is "pseudo-bulk" analysis with RNA-seq methods like edgeR or DESeq2, where "cell type" is encoded as discrete categories. Biologically, discrete categories are not always the most appropriate concept.(1/3)
doi.org/10.1038/s415...

New brain aging study is out today in Nature
The largest single-cell RNA seq dataset of mouse brain aging reveals incredible insights and could pave the way for future therapies to slow or manage the impacts of the aging process. 🧵 #studyBRAIN
Method of moments framework for differential expression analysis of single-cell RNA sequencing data: Cell
www.cell.com/cell/fullte...
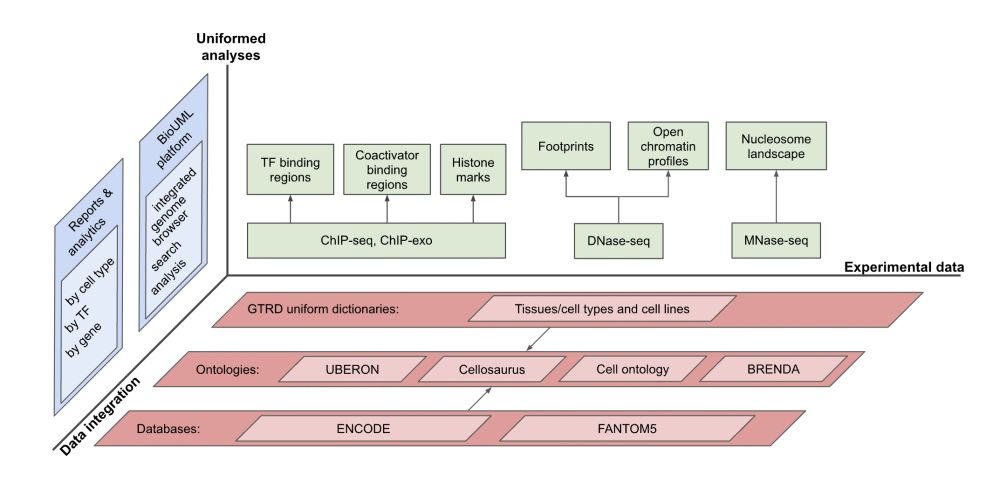
The most complete collection of uniformly processed ChIP-seq data on identification of transcription factor binding sites for human and mouse. Convenient web interface with advanced search, browsing and genome browser based on the BioUML platform. gtrd.biouml.org/
20.12.2024 18:00 — 👍 13 🔁 5 💬 0 📌 0ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements www.nature.com/articles/s4...
19.12.2024 15:15 — 👍 3 🔁 1 💬 0 📌 0Totally agree, real science is really hard! - So much so, that if you don't have fun while doing it, you may get easily frustrated between the "trial and error circuit"
20.12.2024 05:38 — 👍 0 🔁 0 💬 1 📌 0Real science:
1. Is not correlated with the amount of funding
2. Progresses slowly, usually taking many years
3. Leads to more questions than it answers
4. Advances when we disengage & in improvisational discussions
5. Is too important a thing to be done in a non-playful way
🌟 Thrilled to announce that our project, Deep Immune Receptor Modeling (#DIRM), has been awarded funding under the (@novo-nordisk.bsky.social) Nordisk Foundation Data Science Collaborative Research Programme! 🎉
19.12.2024 16:04 — 👍 6 🔁 1 💬 1 📌 0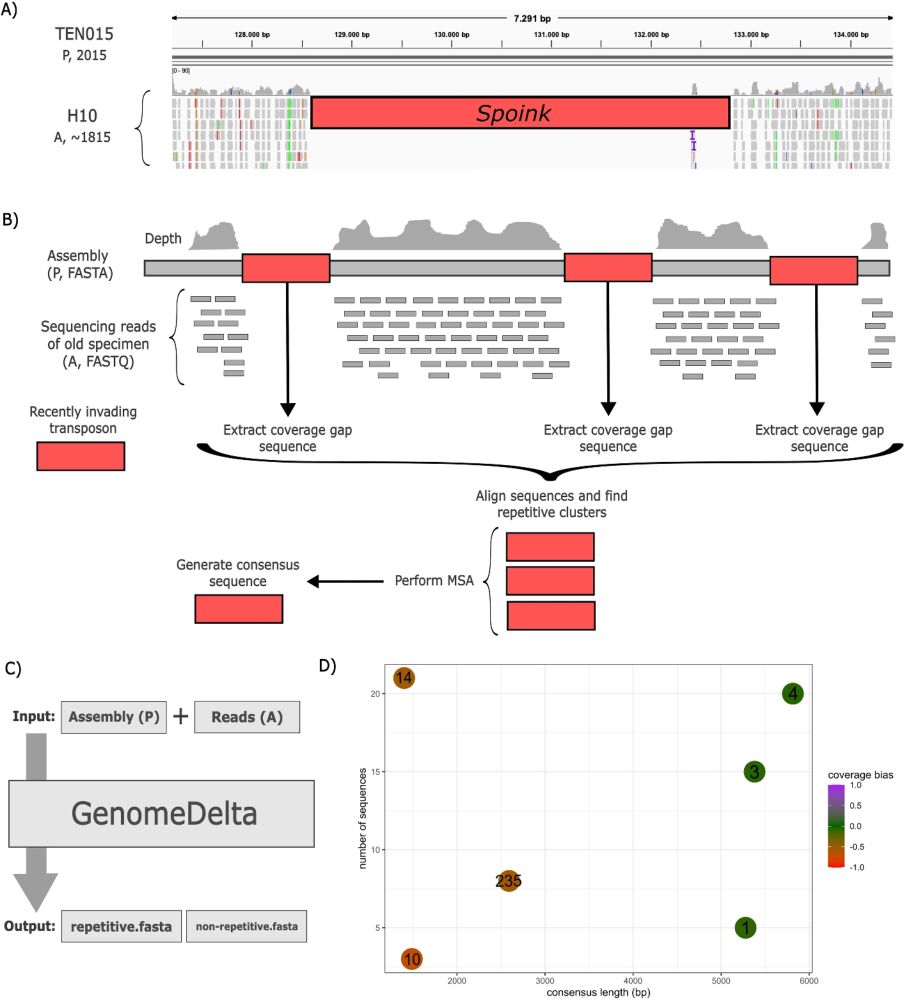
Our novel tool for finding recent TE invasions without repeat library - great work by amazing student Riccardo (not yet on bsky) genomebiology.biomedcentral.com/articles/10....
19.12.2024 08:24 — 👍 4 🔁 5 💬 0 📌 0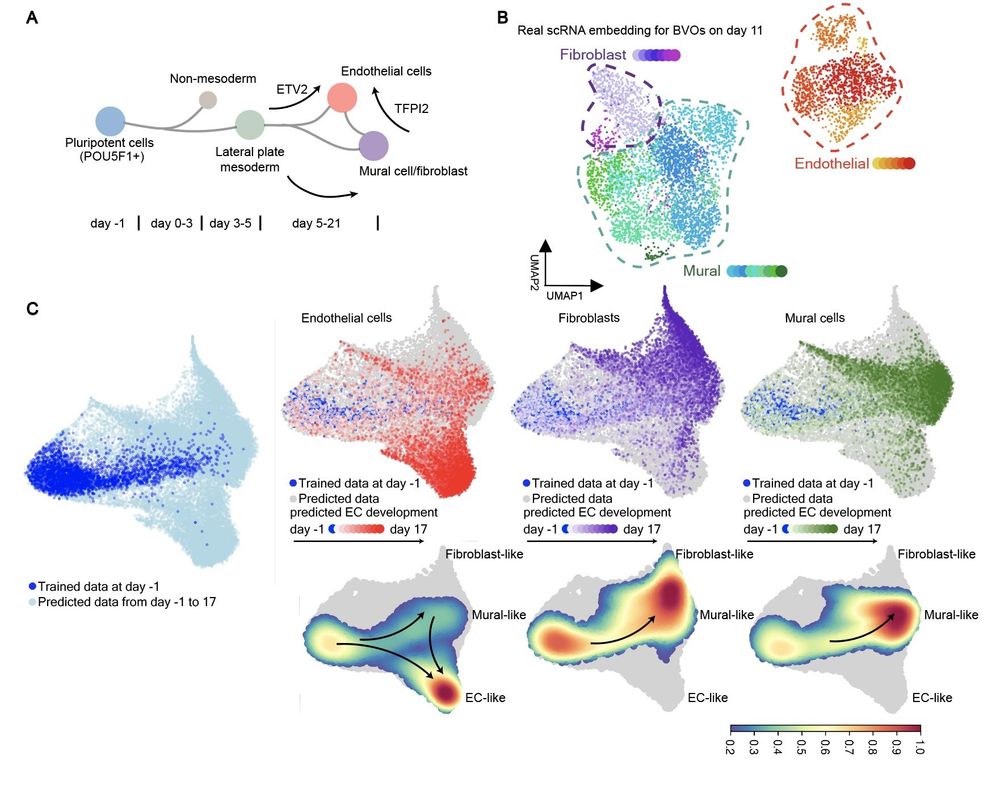
We applied Squidiff to model blood vessel #organoids using single-cell RNA sequencing to examine their response to neutron irradiation and pro-regenerative G-CSF treatment, simulating cosmic irradiation anticipated in #deepspace missions🧑🚀. 5/🦑
09.12.2024 14:13 — 👍 2 🔁 2 💬 1 📌 0Proud of this work spearheaded by the phenomenal @jlfan.bsky.social and @mingxz.bsky.social in collaboration w/ Ben Izar! The past 3 years we've worked hard to unravel how #CNVs shape #tumor phenotypic plasticity seen in #singlecell #RNAseq data ➡️ #Echidna 🦔
18.12.2024 14:08 — 👍 17 🔁 6 💬 1 📌 0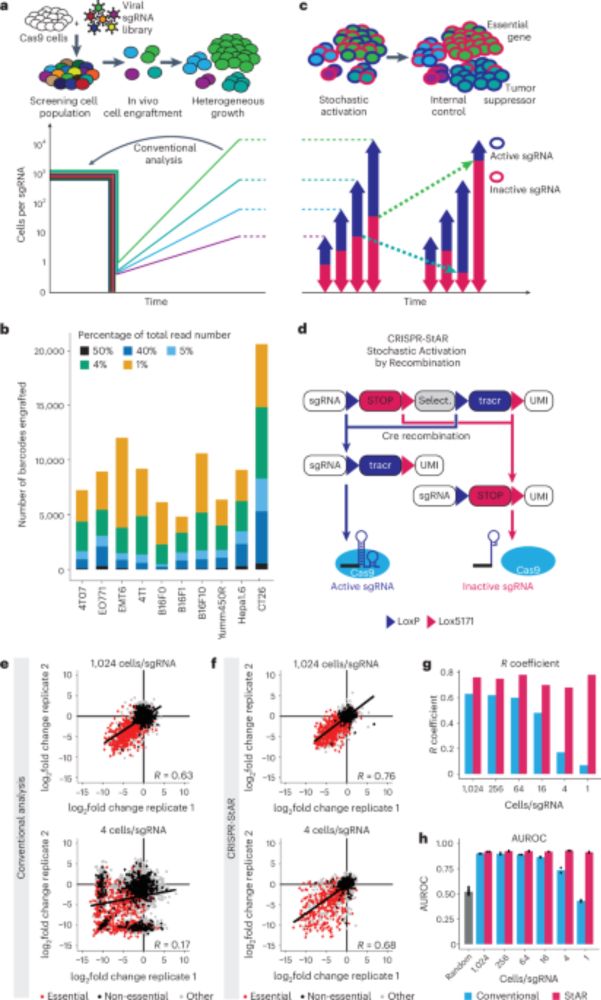
Today we published a new paradigm to do CRISPR screens in complex model systems such as in vivo but also e.g. in organoids. The publication came out in Nature Biotechnology.
1/
www.nature.com/articles/s41...
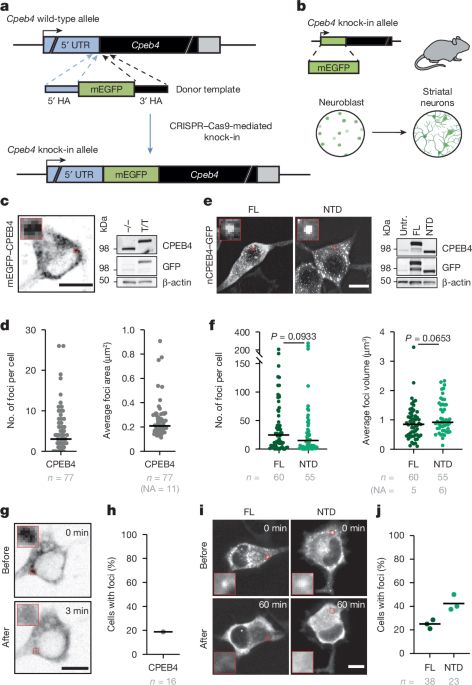
This is huge. Mis-splicing of a 24nt-long micro-exon in the CPEB4 #RNA binding protein leads to formation of irreversible aggregates, which normally would dissolve after depolarization, and consequent deregulation of genes associated with autism spectrum disorders www.nature.com/articles/s41...
09.12.2024 00:13 — 👍 40 🔁 11 💬 0 📌 2