
🚨New preprint out!
🧬Short reads can now decode centromeres.
🌍We reveal population-scale centromere haplogroups and their links to disease.
biorxiv.org/content/10.1... (1/n)
@rainosma.bsky.social

🚨New preprint out!
🧬Short reads can now decode centromeres.
🌍We reveal population-scale centromere haplogroups and their links to disease.
biorxiv.org/content/10.1... (1/n)
If you have questions about our methods or findings, we welcome your feedback and inquiries.
t.ly/OA9fS
Thank you for your interest in our work! (8/n)

Our classifier is freely accessible on GitHub and can be applied to your datasets for KEAP1-NRF2 pathway analysis.
t.ly/S3JUv


When applied to #SRA data, the classifier detected constitutive pathway activation caused not only by known KEAP1-NRF2 alterations in cell lines, but also by previously unreported alterations on the system. (6/n)
13.12.2024 04:48 — 👍 0 🔁 0 💬 1 📌 0
The final model was cross-validated, achieving a
Specificity of 96.90%, discovering as well new alterations beyond the original scope of analysis within #TCGA. (5/n)

Our preliminary model helped reclassify ambiguous mutations, enriching our training data.
This iterative approach ensured diverse mutation representation, improving model robustness. (4/n)


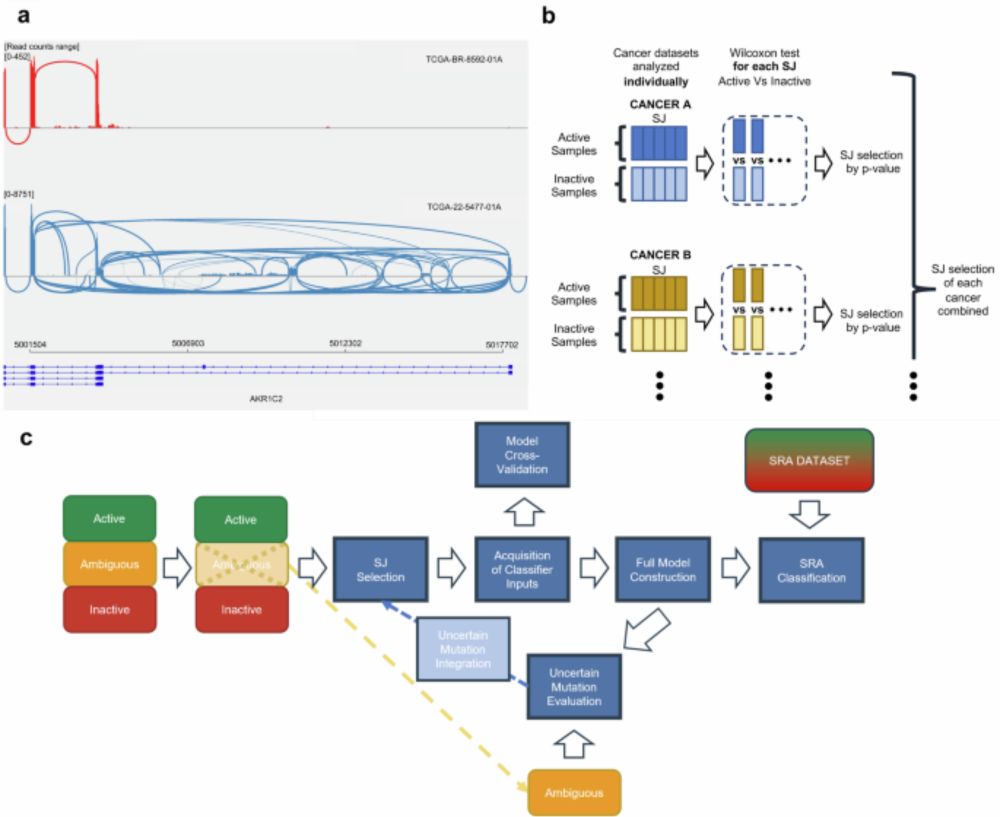
We developed a naive Bayes-based classifier trained on #TCGA data to identify constitutive activation of the KEAP1-NRF2 pathway.
It focuses on mutations impacting pathway activity and leverages splicing junction signatures as biomarkers. (3/n)

The KEAP1-NRF2 pathway plays a critical role in cellular stress responses. Its constitutive activation is linked to genomic alterations, often found in cancer.
We explored a novel way to detect these activations using aberrant splicing junctions. (2/n)

Excited to announce that our paper on the use of splicing junction for detecting abnormal constitutive activation of the KEAP1-NRF2 system, led by @friend1ws.bsky.social at NCCRI, is now published in @natureportfolio.bsky.social npj Systems Biology and Applications! (1/n)
t.ly/OA9fS