Thanks so much Tim!
09.01.2025 06:40 — 👍 2 🔁 0 💬 0 📌 0
Thanks so much Jacob!
08.01.2025 16:08 — 👍 2 🔁 0 💬 0 📌 0
Thank you Alex!!
08.01.2025 05:33 — 👍 1 🔁 0 💬 0 📌 0
I first had this idea 6.5 years ago, early on in grad school. This journey has been a long one. I couldn't have done it without the support and guidance from @genologos.bsky.social and Barak Cohen, our collaboration with @corbolab.bsky.social, or help with the modeling and analysis from my coauthors
08.01.2025 00:18 — 👍 4 🔁 0 💬 1 📌 0
Our solution is to let your model guide you. By focusing on uncertain sequences and testing them functional genomic assays, you can *iteratively* train a model. We applied this to understand why the same DNA sequence motif has radically different effects in different contexts.
08.01.2025 00:18 — 👍 6 🔁 0 💬 1 📌 0
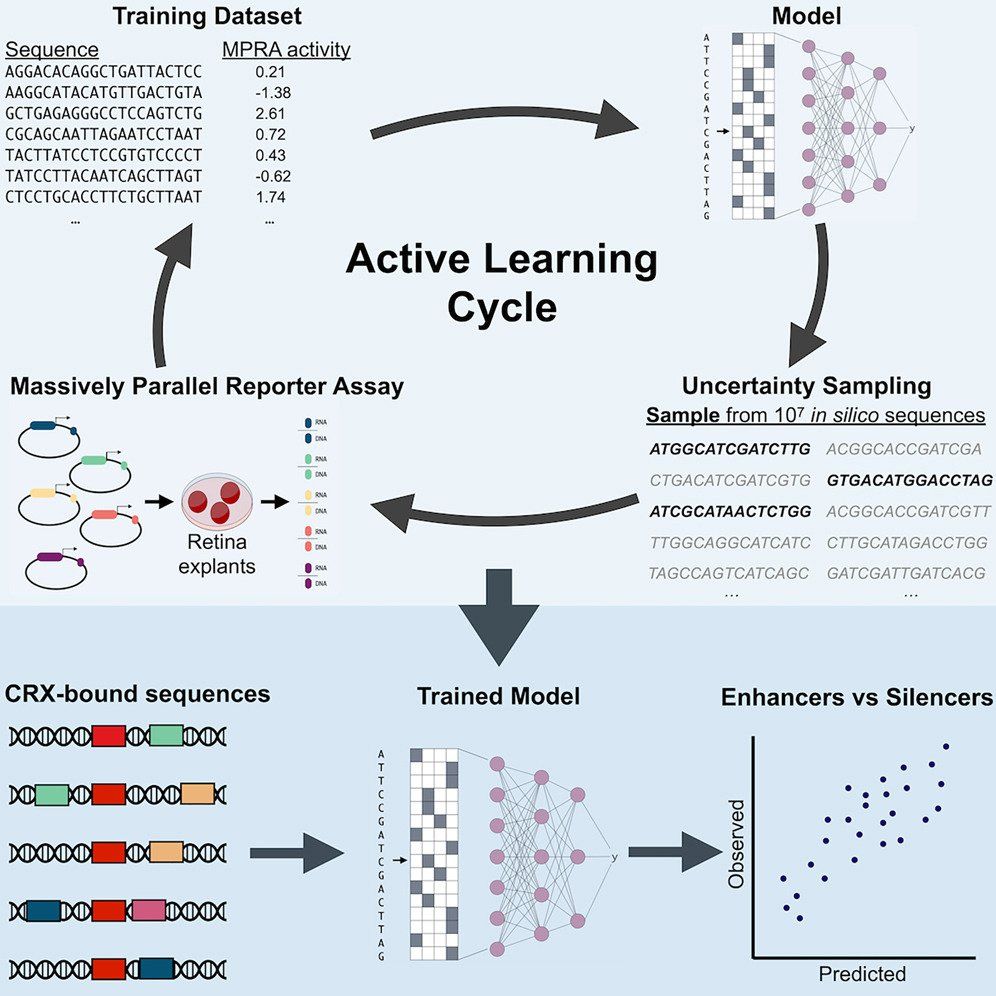
My thesis work on active machine learning to model regulatory DNA is now out in Cell Systems!
We answer the question: When you can synthesize any DNA sequence you want, how do you decide which ones are worth testing?
www.sciencedirect.com/science/arti...
08.01.2025 00:18 — 👍 63 🔁 15 💬 3 📌 4
Many thanks to Yawei Wu, Lloyd Tripp, and Daniel Lyon for their help with these analyses!
The manuscript itself is also restructured. Figs 2 and 4 are swapped, there's a 5th fig for the K562 analysis, and we reworked the Discussion.
Apologies if threading isn't the way to go on Bsky. 🧬🔄
8/8
20.02.2024 19:00 — 👍 2 🔁 0 💬 0 📌 0

We analyzed a second pair of sequences with similar motif content. The model correctly predicts that the RORB motif must be 3' of the CRX motif.
These results show our model learns the context that distinguishes functionally non-equivalent motifs.
7/
20.02.2024 18:59 — 👍 1 🔁 0 💬 1 📌 0

RORB motifs have a wide range of effects when mutated. Our model predicts this correctly & these effects are correlated with motif affinity.
Along with our other results, this shows active learning generates the data needed to learn regulatory grammars.
6/
20.02.2024 18:58 — 👍 1 🔁 0 💬 1 📌 0

We have a new result showing that our model accurately predicts when CRX motifs increase vs. decrease expression. This is crucial because nc variants can change activity in unexpected directions, so it's important to have data that can tell when a motif has a positive vs negative effect.
5/
20.02.2024 18:58 — 👍 1 🔁 0 💬 1 📌 0
Our experiments suggest that inactive sequences are low-information training examples. This is important because large libraries derived from random DNA are mostly inactive seqs. We think iteratively training models on smaller but more informative training data is more effective
4/
20.02.2024 18:57 — 👍 1 🔁 0 💬 1 📌 0


When we did many rounds, active learning was more efficient, approached the upper bound with less data, and enriched for positive examples!
This demonstrates that active learning is broadly effective and illustrate that enriching for active sequences is more informative
3/
20.02.2024 18:57 — 👍 1 🔁 0 💬 0 📌 0


We tested active learning in a second system using Nadav Ahituv and @jshendure.bsky.social's genome-wide MPRA in K562s. We downsampled the data, trained a CNN, then sampled from the remaining data. Active learning consistently outperformed random sampling across many starting conditions.
2/
20.02.2024 18:57 — 👍 1 🔁 0 💬 2 📌 0
We substantially revised our active learning manuscript. A brief summary of what's new.
TLDR: several new analyses, benchmarking w a 2nd MPRA dataset, and a refocused argument on active learning to leverage the capacity of MPRAs to generate large datasets.
www.biorxiv.org/content/10.1...
1/8
🧬🔄
20.02.2024 18:55 — 👍 5 🔁 2 💬 1 📌 0
A project I contributed to during grad school is now up at PLoS Comp Bio! We used the MAVE-NN package by @jbkinney.bsky.social's group to learn about the behavior of synthetic regulatory elements 🧬🔄 Keep an eye on @genologos.bsky.social's Twitter for more details.
journals.plos.org/ploscompbiol...
17.01.2024 01:02 — 👍 0 🔁 0 💬 0 📌 0
I should note that I set up my folders somewhere around my third year of grad school and haven't meaningfully reorganized it since then, so I'm open to a complete overhaul.
17.01.2024 00:55 — 👍 0 🔁 0 💬 0 📌 0
I want to reorganize my @paperpile.bsky.social. I have folders covering broad topics with very few subfolders. Tags are for different manuscripts.
Is there anyone in genomics 🖥️🧬 who wants to share how they organize their reference managers? I'm looking for more meaningful and detailed categories.
17.01.2024 00:55 — 👍 2 🔁 0 💬 1 📌 0
Next time I meet a techie asking how to move into compbio I’ll connect them with you!
05.12.2023 16:30 — 👍 0 🔁 0 💬 0 📌 0
Yeah, I struggle with finding a polite way to say “go learn a bunch of biology or take a huge pay cut to be an entry level bioinformatician for a few years” but there is definitely a mode of thought among some techies that they can watch 20 hours of YouTube videos and call it good
05.12.2023 16:07 — 👍 3 🔁 0 💬 1 📌 0

🤗
05.12.2023 03:56 — 👍 13 🔁 4 💬 0 📌 0
I've had several software engineers ask me recently about transitioning into genomics/comp bio from tech. I never know what to say -- everyone I know in the field in industry went to school for biology/comp bio. Anyone have any suggestions, either of what to say or concrete resources to provide? 🧬🖥️
04.12.2023 22:21 — 👍 5 🔁 1 💬 4 📌 0
Agreed that there is strong cell to cell variability. But changes in e.g. the ZRS enhancer of Shh can cause loss of limbs or gain of extra digits. Some of that is due to changes in Shh in space/time, but some is also due to how *much* Shh is produced
11.10.2023 21:21 — 👍 1 🔁 0 💬 1 📌 0
hmm...A difference in 2 vs 3 fold change can definitely matter! Over/underactive CREs can cause developmental defects and disease. Agreed that cell culture won't tell you space+time, but they can tell you information about how sequence features encode activity.
11.10.2023 21:13 — 👍 1 🔁 0 💬 1 📌 0
My expertise is also in distal elements, but I think that diffs in DBDs reflects different points in evolution. TFIIB and the sigma factors are homologous. But most DBD families in euks are totally absent from proks and vice versa! Suggests txn initiation is more conserved than specifying space-time
11.10.2023 21:12 — 👍 1 🔁 0 💬 0 📌 0
I'm not entirely sure, but my sense is that GTFs have structural classes of DBDs you don't see binding to distal regions
11.10.2023 20:43 — 👍 2 🔁 0 💬 1 📌 0
I think that enhancers+silencers control space and time as well as expression levels 😊
11.10.2023 20:39 — 👍 1 🔁 0 💬 1 📌 0
I think of promoters as regions where transcription is initiated, while enhancers (and silencers!) are regions that dictate how much RNA is produced.
11.10.2023 19:29 — 👍 2 🔁 0 💬 1 📌 0
I think it depends what counts as a promoter. If you think that the promoter is only the ~100 bp core promoter near the TSS, then there is a difference because the core is bound by GTFs. If you consider the ~1kb upstream of the promoter as the promoter, more overlap.
11.10.2023 19:29 — 👍 2 🔁 0 💬 2 📌 0
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Assistant Professor at UW studying, evo-devo, annual killifish, & cell type evolution.
PhD Student @ UW Genome Sciences
Population geneticist interested in evolutionary consequences of infectious disease for both hosts and pathogens, and everything single-cell!
Postdoctoral Fellow in the Department of Genome Sciences at the University Washington
Award-winning home tank for all things zebrafish! Learn more about us and our Slack tankspace at our website: linktr.ee/zebrafishrock
Director Duke Sequencing & Genomics Tech / Asst Prof MGM / cis-regulatory evolution / DEI / Mama / she | her / daughter of immigrants / wearer of many hats, owner of one head / posts my own
PhD student at Cohen Lab, WashU | gene regulation, genomics🧐
It's like Gmail for your papers - a modern reference manager.
We love papers and post about publishing, academic productivity and everything related.
Professional sceptic; never heard an uninteresting fact about T cells; I like microorganisms/microbiomes too.
Microbiome research, microbial (meta)genomics, bioinformatics
Bioinformatics Scientist at CHOP Microbiome Center
✝️she/her
Lefty in tech. Trying to push for progress and enjoy what I can
🏳️🌈| he/him
PhD candidate at UW Seattle. I study how interactions between viruses shapes their evolution. Also 📷, 🎹, 📽️, 🐦
Physician-scientist in-training and hyphen-enthusiast interested in hematology, aging, and somatic mosaicism
Banting Postdoc with @JShendure @uwgenome | Prev
@CIHR_IRSC UBC Neuro PhD | Genome engineering, molecular recording, neurodevelopment and its disorders | Mountains & skateboarding
PhD Candidate @ UBC. Genomics, Gene regulation and Data Science @ de Boer Lab.








