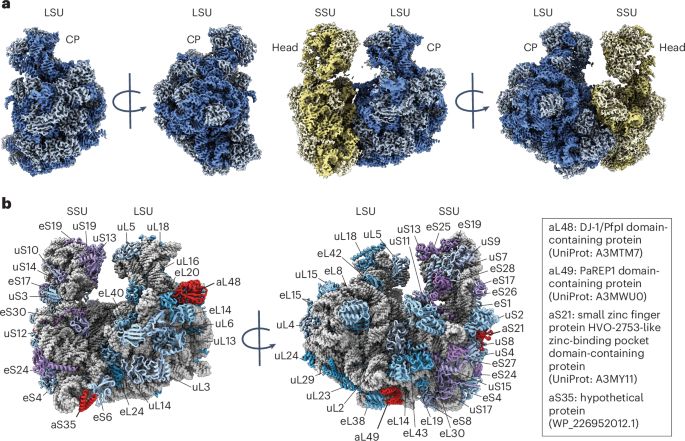
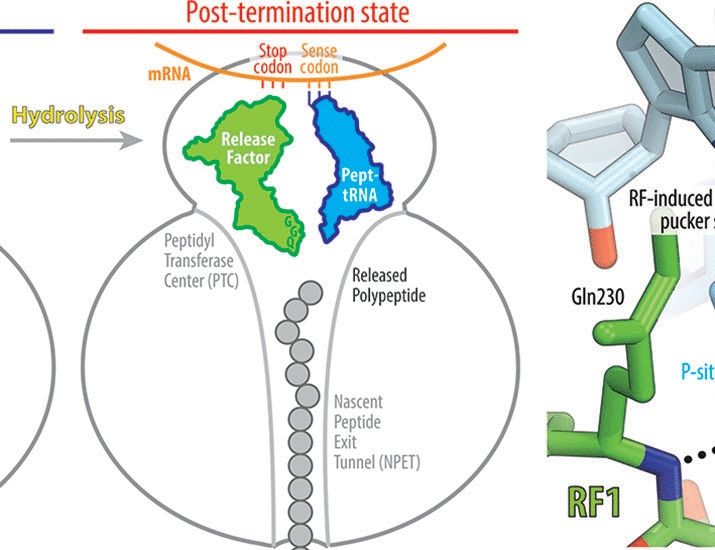
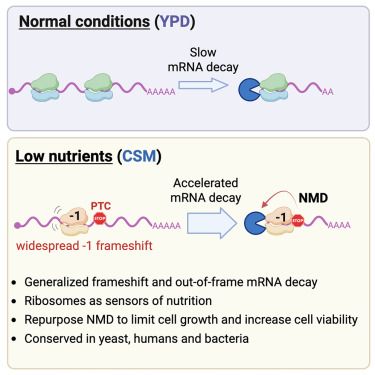
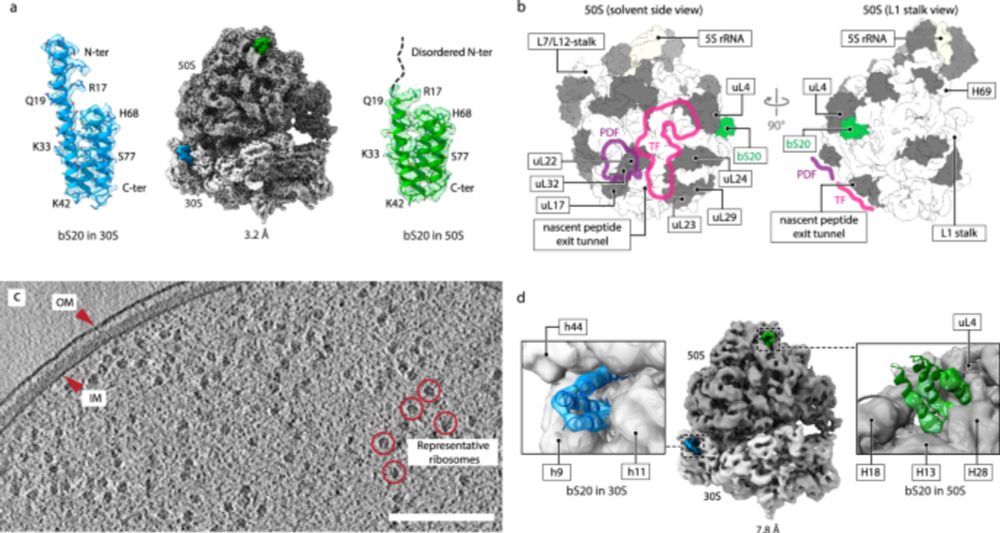
Thank you zeynep. It’s very interesting and useful. 👍
16.09.2025 06:59 — 👍 1 🔁 0 💬 1 📌 0
#RNAsky #RNAbiology
12.09.2025 19:23 — 👍 1 🔁 2 💬 0 📌 0

Translon: a single term for translated regions - Nature Methods
Nature Methods - Translon: a single term for translated regions
A large consortium of researchers introduces the term 'translon' to denote any transcriptome region that is decoded by ribosomes.
www.nature.com/articles/s41...
05.09.2025 13:30 — 👍 21 🔁 6 💬 1 📌 2
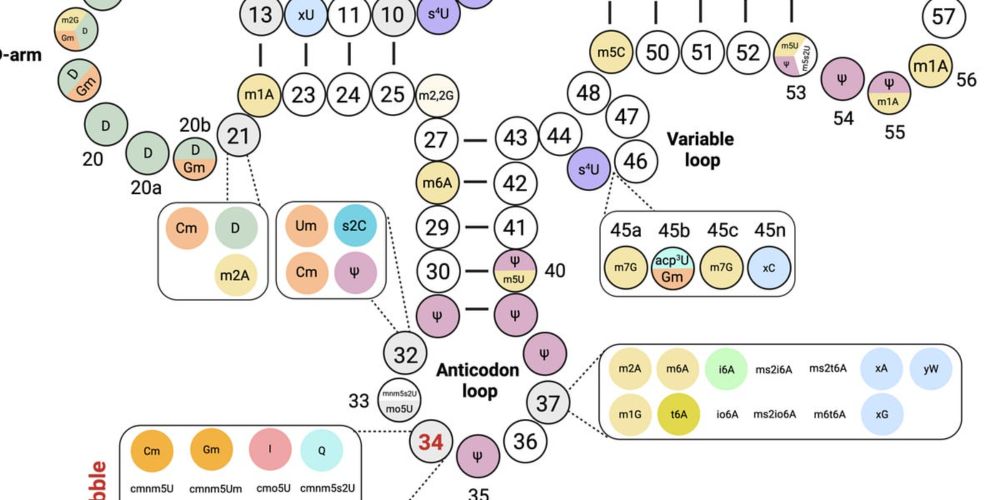
RNA modifications digest Summer 2025
Bimonthly newsletter: RNA modifications and related topics, mostly in bacteria. Summer 2025, first issue!
🦠 Launching a bimonthly digest on RNA modifications and related topics in bacteria!🤩 First issue: Summer 2025. Shared here + by email, future ones will be quicker reads! 😅 #rnasky #microsky #ribosome Subscribe and share if you are interested! rnamodifupdates.substack.com/p/rna-modifi...
15.08.2025 09:30 — 👍 17 🔁 11 💬 0 📌 0
Frameshifting Stimulatory Sequence Induces Large Structural Change of Ribosomal Proteins When Bound to E. coli Ribosomes pubmed.ncbi.nlm.nih.gov/40788995/ #cryoEM
12.08.2025 18:40 — 👍 1 🔁 1 💬 0 📌 0
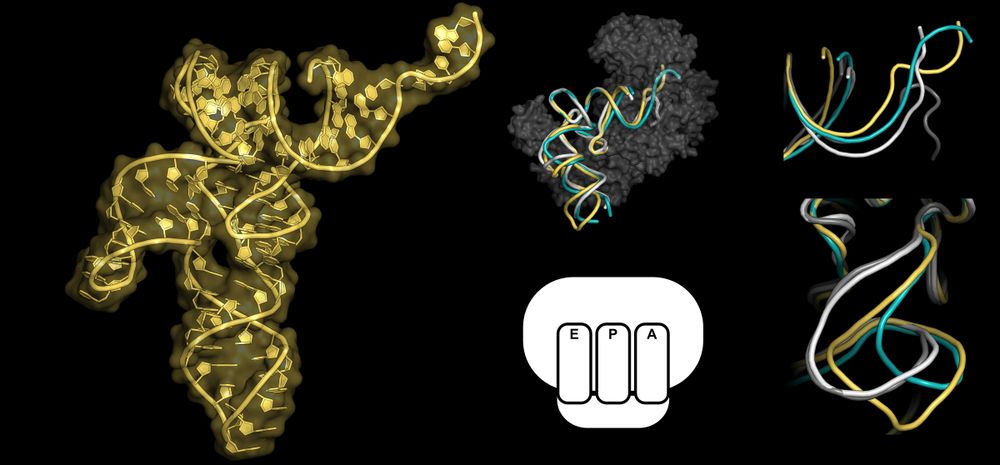
Free tRNALeu shows flexible variable arm and CCA end, reshaped by LeuRS and ribosome binding #tRNA #StructuralBiology bit.ly/45PxJF5
31.07.2025 13:51 — 👍 7 🔁 3 💬 0 📌 1
Finally out!
I’m thrilled to share our new paper (Wolin et al., Cell 2025).
This paper describes SPIDR, a high-throughput method for mapping RBP binding sites.
By combining #SPIDR with #cryoEM, we identified the exact binding site of LARP1 within the #mRNA channel of the 40S ribosomal subunit.
22.07.2025 17:12 — 👍 66 🔁 21 💬 4 📌 1
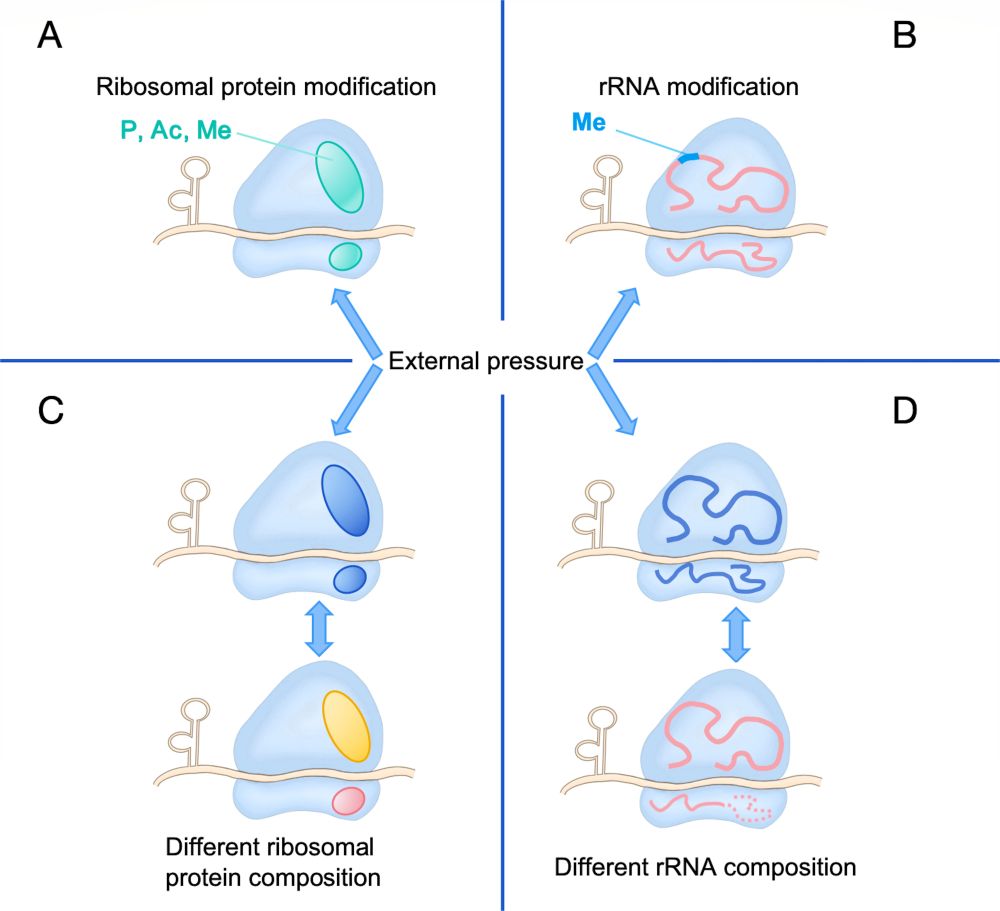
Editor's Pick: Shen, Ye et al. review recent literature on the role of bacterial ribosome heterogeneity on facilitating rapid response to stress.
journals.asm.org/doi/10.1128/...
@asm.org #JBacteriology
21.07.2025 14:37 — 👍 33 🔁 18 💬 1 📌 2
Insights on how ribosomal RNA modification distant from drug binding site can still affect drug binding, in mycobacterium 🦠 #rnasky #ribosome #microsky
10.07.2025 07:33 — 👍 6 🔁 3 💬 0 📌 0

The cover of the April issue of @cp-trendspharma.bsky.social. Gregory Howard and William Tansey review the current status, challenges, and future prospects of cancer treatments that target the ribosome. Aberrant ribosome production and function are hallmarks of cancer.
22.04.2025 16:36 — 👍 13 🔁 8 💬 1 📌 0

Nouvelle facette de la protéine Rqc2, sous-unité d’un système de sauvetage des ribosomes chez Saccharomyces cerevisiae
Les cellules utilisent de nombreux mécanismes de contrôle-qualité au cours de la biosynthèse des protéines. Les défauts qui perturbent le décodage de l'ARN messager peuvent provoquer le blocage des ribosomes, empêchant l'expression des gènes. Ces ribosomes bloqués ne sont plus disponibles pour le processus de traduction, ce qui représente un défi à la fois pour l'homéostasie des ribosomes et la survie de la cellule.
Les cellules eucaryotes évitent l'accumulation de polypeptides aberrants potentiellement toxiques et maintiennent la disponibilité des ribosomes en employant des mécanismes de surveillance, comme le complexe de contrôle-qualité associé aux ribosomes (RQC), très conservé au cours de l'évolution. Les voies de sauvetage dissocient le ribosome bloqué, en libérant la sous-unité 40S attachée à l'ARNm et la sous-unité 60S encore attachée à un ARNt avec le peptide en élongation. RQC assure l'ubiquitination, l'extraction et la dégradation du peptide naissant aberrant. En particulier, Rqc2p recrute, à la même position que l’ARNt au site A, des ARNt chargés en alanine et en thréonine, formant des extensions C-terminales composées de ces résidus (CAT-tails). Ces CAT-tails peuvent exposer des résidus lysine cachés dans le tunnel de sortie de la 60S pour l’ubiquitination par la E3 ligase Ltn1.
En utilisant un crible génétique, les scientifiques de l’équipe Génomique, Structure et Traduction à l’I2BC (CNRS/CEA/UPSaclay, Gif-sur-Yvette) ont identifié un allèle mutant de RQC2 comme impliqué dans la libération des peptides à partir des ribosomes bloqués. Ils ont caractérisé la protéine mutante sur le plan biochimique et, en collaboration avec l’équipe de Reynald Gillet (IGDR, Université de Rennes), sur le plan structurel, en déterminant comment la mutation ponctuelle réduisait le recrutement de l'ARNt au site A, limitant ainsi la formation des CAT-tails.
Ces résultats, publiés dans Structure, permettent de mieux comprendre le rôle de Rqc2p dans le complexe de contrôle-qualité associé aux ribosomes, et le maintien de l'homéostasie des ribosomes.
-> Contact : celine.fabret@i2bc.paris-saclay.fr
Nouvelle facette de la protéine Rqc2, sous-unité d’un système de sauvetage des ribosomes chez Saccharomyces cerevisiae
15.04.2025 19:41 — 👍 2 🔁 1 💬 0 📌 0
This work has been done at #I2BC @cnrsbiologie.bsky.social @cnrs-paris-saclay.bsky.social @i2bcparissaclay.bsky.social
06.04.2025 19:43 — 👍 2 🔁 1 💬 0 📌 0
Using cryoEM we observed This mutation promotes a misplacement of tRNA in the 60S altering the formation of #CATtails.
06.04.2025 19:41 — 👍 1 🔁 0 💬 1 📌 0
We performed a genetic screen to identify factor involved in the release of peptide from stalled #ribosomes. We identified a new mutation in #RQC2 factor.
06.04.2025 19:41 — 👍 0 🔁 0 💬 1 📌 0
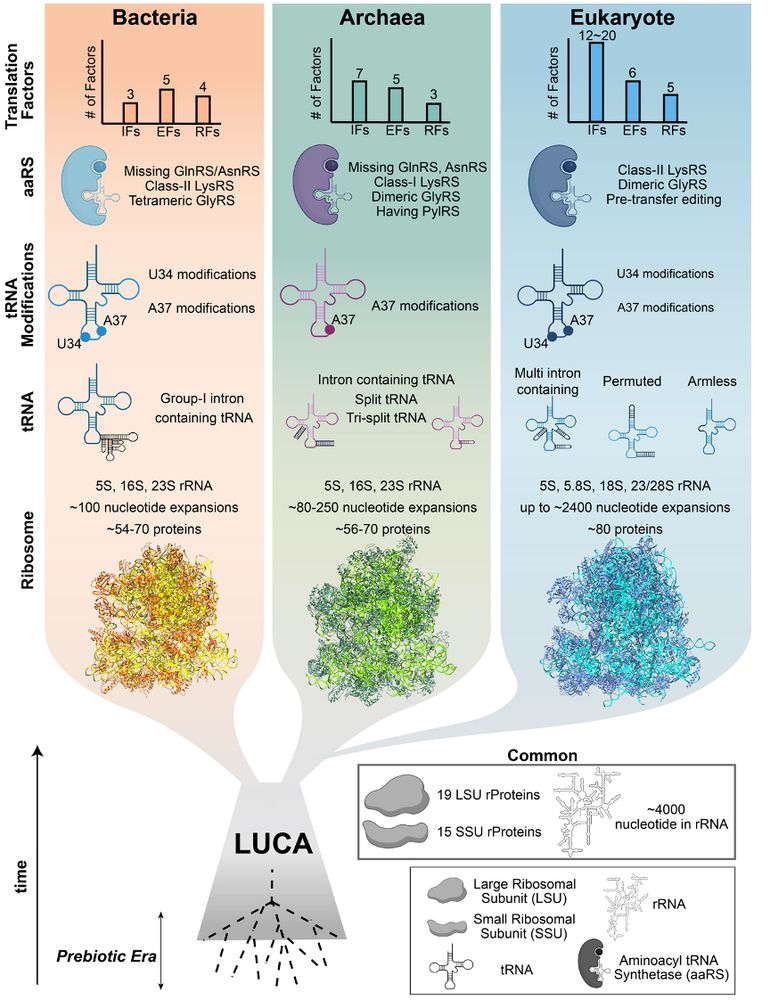
🚨New paper!🚨
A comprehensive take on the origins and evolution of translation factors & how these essential players evolved across the tree of life. 🌍🧬
Led by Evrim Fer @uwmadisonmdtp.bsky.social grad student! 👏
Free access: www.sciencedirect.com/science/arti... @cp-trendsgenetics.bsky.social
24.03.2025 14:46 — 👍 94 🔁 46 💬 3 📌 4
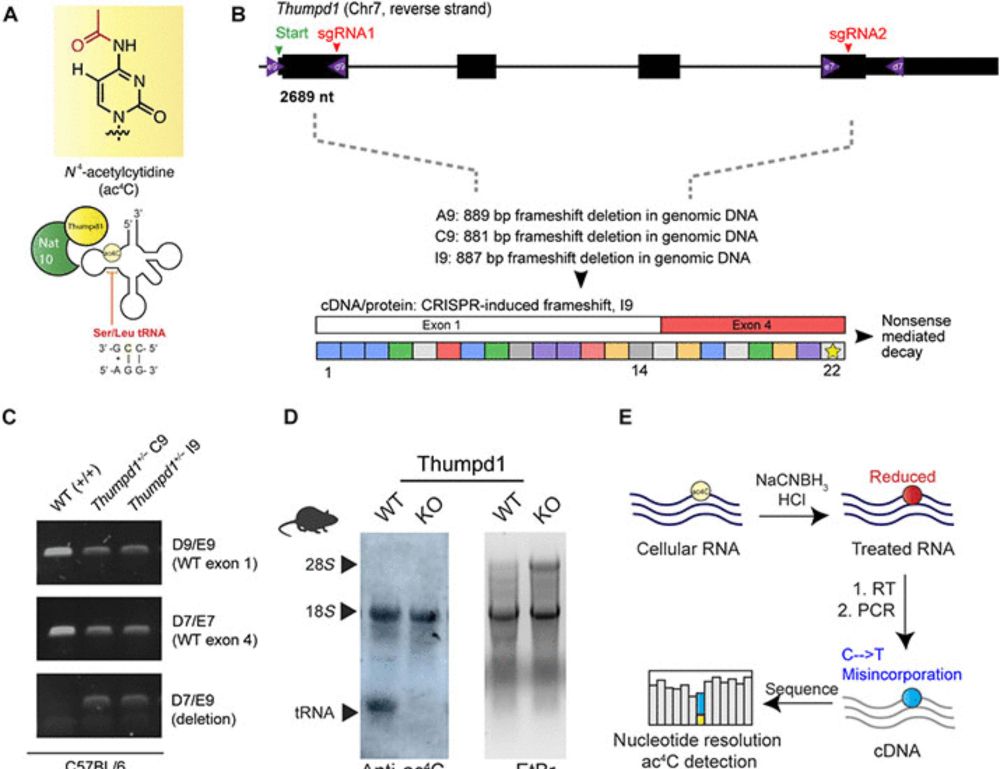
Transfer RNA acetylation regulates in vivo mammalian stress signaling
An ancient tRNA modification is used by mammalian cells to coordinate protein translation and adaptive signaling.
Excited to share this one! We developed an in vivo model for specific manipulation of transfer RNA acetylation and found it serves as a sentinel modification whose loss causes ribosome stalling and stress signaling. Implications for a genetic disorder and cancer.
www.science.org/doi/10.1126/...
20.03.2025 01:42 — 👍 101 🔁 27 💬 8 📌 3
RNA biologist at Johannes Gutenberg University Mainz, Germany: co-translational quality control - RNA modification - and beyond.
Lab url: ak-winz.pharmazie.uni-mainz.de
Scientist studying ribosome stalling and translatome remodelling in fission yeast at the Department of Biochemistry, University of Cambridge
Postdoc at the Max Planck Institute for Multidisciplinary Sciences. Former MSC fellow. Interested in ribosome, translation, ncRNA, DNA replication, microbiology.
PhD student in @balikovalab.bsky.social at @czechacademy.bsky.social of Sciences, BIOCEV, @charlesuni.cuni.cz in Prague
💊 antibiotics 💎 ribosome
🧫 resistance 🧬 ABC-Fs
🦠 staph ❄️ cryo-em
@mbuavcr.bsky.social
@sciencecharles.bsky.social
Professor, Weizmann Institute
Director, Moross Integrated Cancer Center
President, European Association for Cancer Research
Cell aims to publish the most exciting and provocative research in biology. Posts by Scientific Editors on the Cell editorial team. See the latest papers at https://www.cell.com/cell/
Molecular Cell is a Cell Press journal that aims to publish the best papers in molecular biology.
Scientific journal publishing research, overview and commentary across all of biology. All of it!
https://www.cell.com/current-biology/home
Part of CellPress @cellpress.bsky.social
Translation mechanisms team @BIOC @CNRS.bsky.social @Ecole Polytechnique @IP-Paris.bsky.social
Archaea Ribosome Evolution CryoEM
L’actualité des laboratoires du CNRS sur le territoire #ParisSaclay, cluster scientifique, technologique et d'innovation.
Scientist. My primary fediverse account is on mastodon: @cosminribo@framapiaf.org
Bsky users can see my posts there by following the, arguably long, @cosminribo.framapiaf.org.ap.brid.gy
Engineer at the bioinformatics platform (BIOI2) of the I2BC
Senior Editor at Molecular Cell
Cell Biologist, Biochemist, Cryo-ET, Cryo-EM
The Gigant/Ménétrey lab at I2BC is interested in investigating the molecular mechanisms that govern intracellular trafficking processes, especially #cytoskeleton regulation, motor-based transport and organelle trafficking.
Assistant professor at Paris-Saclay University & at Institute for integrative biology of the cell (I2BC). Interested in Archaea, hyperthermophiles, DNA topology, topoisomerases.
Group leader in the Genome Biology Dpt of I2BC. My team studies how loss of function mutations in chromatin remodelers lead to cancer.
Associate Professor @YaleNeuro • co-Director of Graduate Studies @Yale_INP • @YaleRNA • RNA Neurobiology • Neurodegenerative disease • Immigrant • 🐶 dad • Alum @WhiteheadInst @HopkinsNeuro @PKU1898 🏳️🌈
Décrypter la complexité et la diversité du vivant
🔗 https://www.insb.cnrs.fr/fr