@jhigginswriter.bsky.social wrote an article about my lab and our frog 🐸 at the Hutch! Thank you!
29.10.2025 06:29 — 👍 16 🔁 5 💬 0 📌 0

Fred Hutchinson Cancer Center
Fred Hutch is dedicated to the elimination of cancer and related diseases as causes of human suffering and death.
Looking for your next step in fly genetics? The Rajan Lab at Fred Hutch is hiring a staff scientist + postdoc for projects on fat–brain signals, immunity, and brain aging. Stable, long-term role in a collaborative lab. Apply: careers-fhcrc.icims.com/jobs/30062/j...
16.09.2025 23:32 — 👍 25 🔁 11 💬 1 📌 0

Fred Hutchinson Cancer Center
Fred Hutch is dedicated to the elimination of cancer and related diseases as causes of human suffering and death.
My awesome Drosophila colleague Akhila Rajan at Fred Hutch (Seattle) is recruiting both a staff scientist and a postdoc to study fat–brain communication, innate immunity, mitochondrial signaling, and brain senescence. Great team, great environment. Apply here: careers-fhcrc.icims.com/jobs/30062/j...
17.09.2025 01:05 — 👍 47 🔁 38 💬 0 📌 1
One more movie!
Using the RL algorithm (from pnas.org/doi/10.1073/...),
@masaashimazoe.bsky.social analyzed our nucleosome tracking data and classified them by their mobility.
Hotter color = faster motion (Slow🔵→ 🟢 → 🟡 → 🟠 Fast)
29.08.2025 00:36 — 👍 46 🔁 16 💬 2 📌 0

WORK!
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍
13.08.2025 13:30 — 👍 23 🔁 30 💬 2 📌 5
The peer-reviewed VOR for the MagIC-cryo-EM paper by @yarimura.bsky.social and @1001hak.bsky.social is finally published!
Congrats Yasu!
20.05.2025 17:55 — 👍 12 🔁 2 💬 1 📌 0

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
06.05.2025 15:08 — 👍 103 🔁 24 💬 4 📌 3
Excited to share our preprint on the molecular architecture of heterochromatin in human cells 🧬🔬w/ @jpkreysing.bsky.social, @johannesbetz.bsky.social,
@marinalusic.bsky.social, Turoňová lab, @hummerlab.bsky.social @becklab.bsky.social @mpibp.bsky.social
🔗 Preprint here tinyurl.com/3a74uanv
11.04.2025 08:35 — 👍 359 🔁 141 💬 12 📌 20
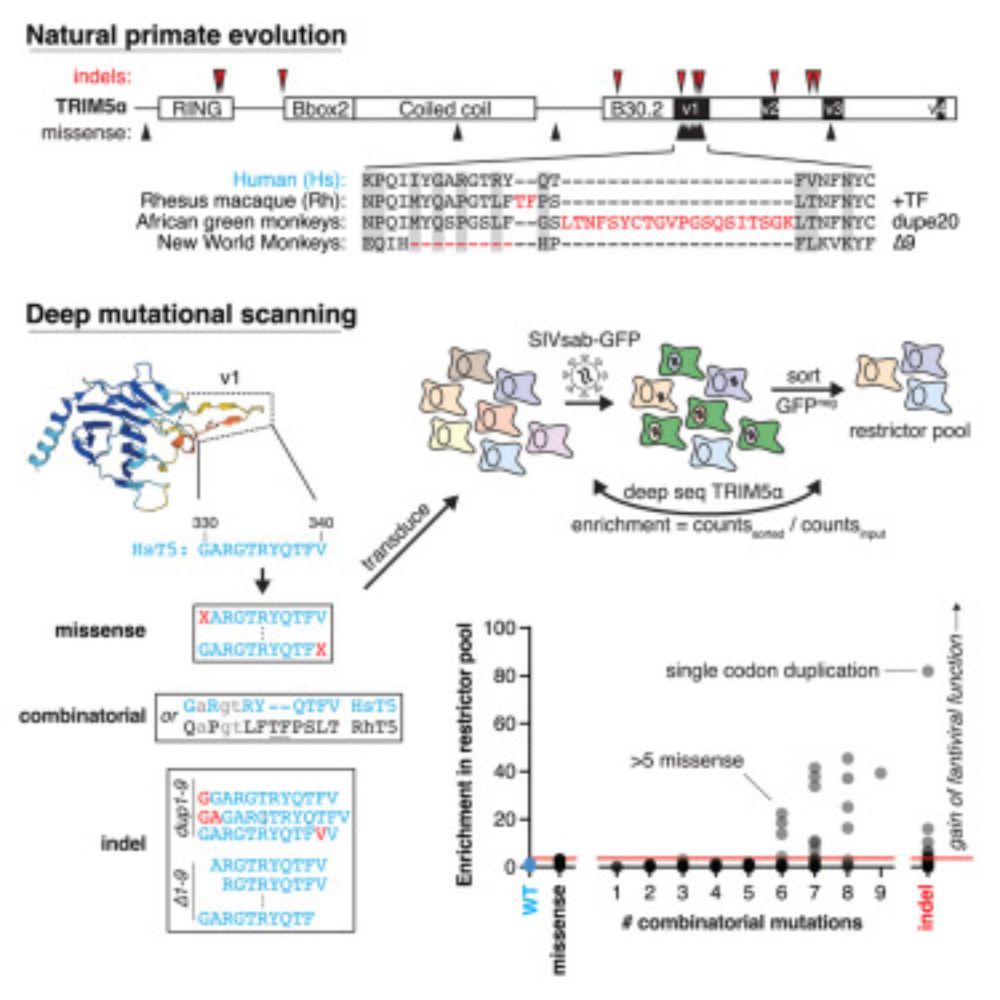
Histone H3 cysteine 110 enhances iron metabolism and modulates replicative life span in Saccharomyces cerevisiae
Histone H3 cysteine 110 enhances iron metabolism and modulates replicative life span in yeast.
Ever wondered what the lonely histone H3C110 does? We found that it’s lost in many fungi. Adding it back boosts histone H3 copper reductase activity, rescues iron defects and modulates lifespan, suggesting an evolutionary tradeoff between these effects.
www.science.org/doi/10.1126/...
11.04.2025 21:43 — 👍 56 🔁 18 💬 1 📌 0
New preprint on 3D heterochromatin architecture in human cells! Great collab with @sergiocruzleon.bsky.social & @johannesbetz.bsky.social from @hummerlab.bsky.social, @marinalusic.bsky.social & the Turoňová lab. Many thanks to my supervisor @becklab.bsky.social. bioRxiv: tinyurl.com/3a74uanv 🧵👇
11.04.2025 09:03 — 👍 224 🔁 67 💬 3 📌 4
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
06.04.2025 09:45 — 👍 179 🔁 57 💬 10 📌 8
Our new paper is out@ScienceAdvances👇
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
29.03.2025 01:06 — 👍 69 🔁 24 💬 1 📌 1
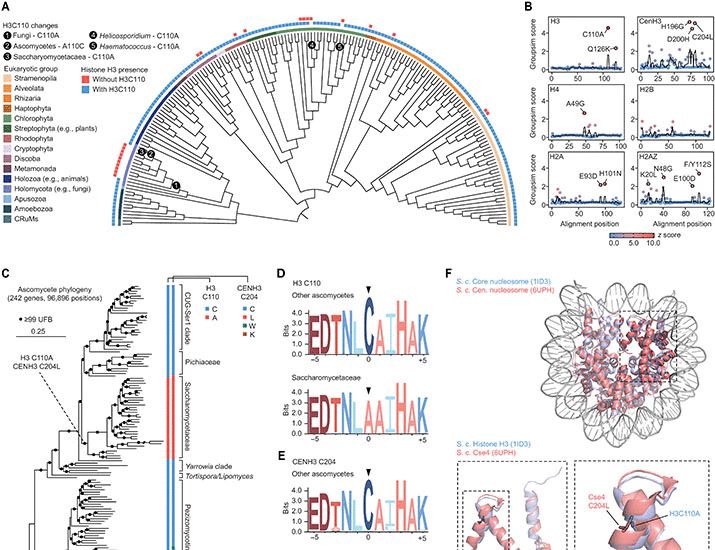
We quantitatively measured nucleosome motion across the 4 chromatin classes, from euchromatin to heterochromatin.
🧪 More euchromatic regions (i.e., earlier-replicated regions) show greater nucleosome motion.
🎉 Congrats @katsuminami.bsky.social @kakonakazato.bsky.social and all collaborators!
2/2
29.03.2025 01:06 — 👍 15 🔁 4 💬 0 📌 0
In a new Science study, cryo–electron tomography captures the in-cell architecture of the mitochondrial respiratory chain, illuminating how the coordinated action of molecular machines drives life’s fundamental energy conversion.
Learn more in this week's issue: scim.ag/3FA3Ygq
20.03.2025 18:05 — 👍 463 🔁 144 💬 15 📌 35
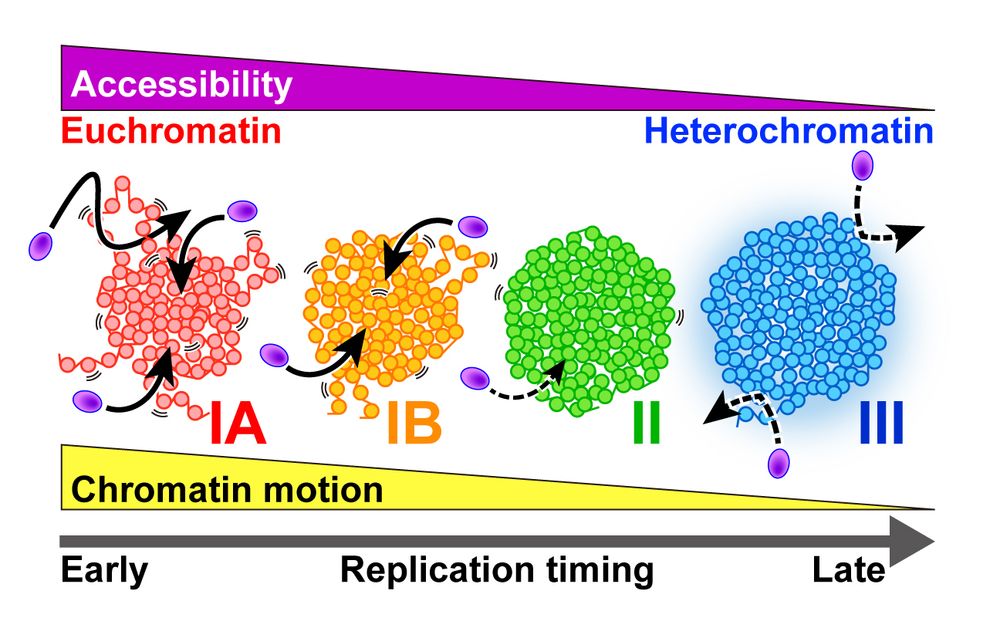
Cryo-ET visualization of chromatin at the nucleosome level in RPE-1 cells arrested in G1 phase and metaphase.
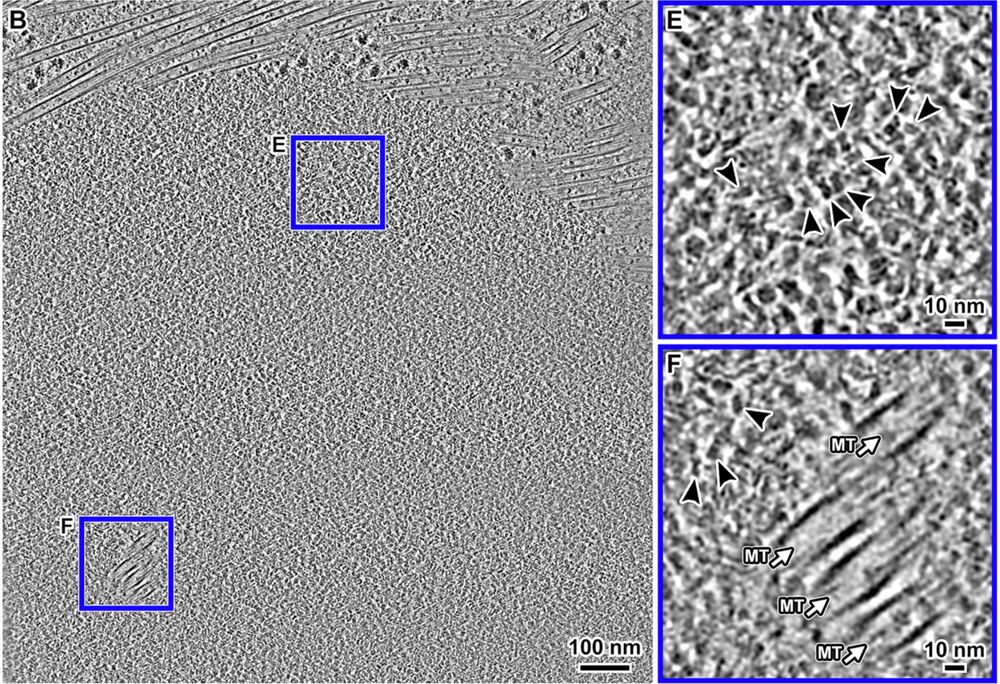
Cryotomograms of a metaphase RPE-1 cell.
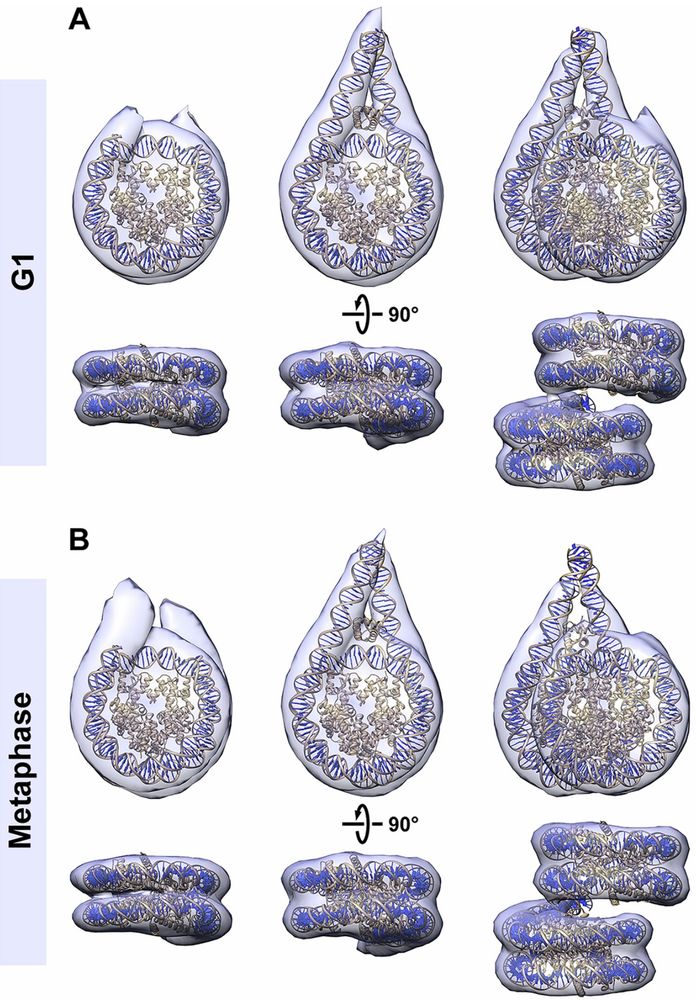
3-D class averages of mononucleosomes and ordered stacked dinucleosomes in G1 and metaphase.
Happy that Jon Chen @jonchenjk.bsky.social's cryo-ET and confocal study of RPE-1 chromatin in situ is online www.embopress.org/doi/full/10.1038/s44318-025-00407-2
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
17.03.2025 21:36 — 👍 22 🔁 11 💬 1 📌 0

Wonderful visit to @hutchbasicsci.bsky.social today and thanks to @yarimura.bsky.social for hosting and dinner with the always entertaining @suebiggins.bsky.social and @harmitmalik.bsky.social. A day discussing science with amazing people is unbeatable.
12.03.2025 05:26 — 👍 44 🔁 4 💬 1 📌 0
Thank you for visiting us and giving the exciting seminar!
12.03.2025 05:31 — 👍 3 🔁 0 💬 0 📌 0
My first, first author paper is now on bioRxiv! 🤩
Linker histone H1 is a liquid-like "glue" condensing chromatin, which revises textbooks! 📖✨
biorxiv.org/content/10.1...
Huge thanks to my PI, @kazu-maeshima.bsky.social, for supervision.
Amazing collab with @rcollepardo.bsky.social’s group!" (1/n)
10.03.2025 09:12 — 👍 17 🔁 6 💬 1 📌 0
I am super excited to share that my work at @yifan_ucsf has just been published in Nature🎉 From developing pYC KI system, it was a long journey to understand endogenous protein dynamics. I appreciate the support from FASN team! 🕺Let's dive into the endogenous & dynamic protein world🏊🏄🚣
02.03.2025 10:25 — 👍 12 🔁 3 💬 1 📌 0

Researchers at Fred Hutch have discovered an overlooked mechanism driving aggressive breast and brain tumors involving genes so ancient — more than 2 billion years old — that they fly under the radar of standard genetic sequencing methods. 1/9 www.science.org/doi/10.1126/...
13.02.2025 19:28 — 👍 36 🔁 15 💬 1 📌 5
Huge congratulations @jbloomlab.bsky.social!
15.02.2025 16:56 — 👍 7 🔁 2 💬 0 📌 0
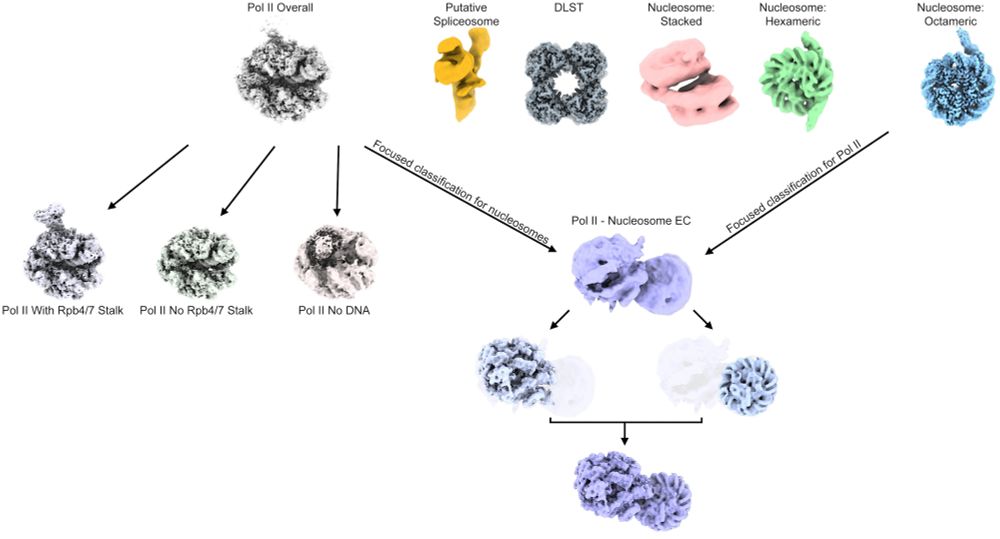
We did a thing... We purified native transcriptional complexes from Drosophila embryos, obtaining +/- stalk Pol II elongation complexes, native nucleosomes, as well as a nucleosome elongation complex. For more details please check out: doi.org/10.1101/2025...
05.02.2025 01:02 — 👍 56 🔁 20 💬 1 📌 1
NOT-OD-24-110: Notice of Legislative Mandates in Effect for FY 2024
NIH Funding Opportunities and Notices in the NIH Guide for Grants and Contracts: Notice of Legislative Mandates in Effect for FY 2024 NOT-OD-24-110. NIH
On changes to #NIH indirect rates, there is a law in place that prohibits NIH from making such changes without the approval of Congress. See Division D, Title II Section 224 of The Further Consolidated Appropriations Act, 2024 (Public Law No: 118-47) grants.nih.gov/grants/guide...
08.02.2025 00:56 — 👍 3610 🔁 1474 💬 78 📌 239
Structural Characterization of Native RNA Polymerase II Transcription Complexes and Nucleosomes in Drosophila melanogaster https://www.biorxiv.org/content/10.1101/2025.02.03.636274v1
04.02.2025 02:20 — 👍 2 🔁 2 💬 0 📌 0
Towards diversity and equity in brain sciences | Become a member: http://alba.network | a division of FENS | Founding Partners @fens.org @ibroorg.bsky.social @sfn.org
Wellcome Trust PhD student in the Philpott Lab at the Cambridge Stem Cell Institute 🔬🧬 Interested in early embryo development, epigenetics & cell fate decisions
Group leader at the Francis Crick Institute. Studying DNA replication one molecule at a time.
Postdoc in the Musacchio Lab @ MPI-Dortmund. Cell division and everything that goes wrong with it. 🇪🇺 He/him
Assistant Investigator at the Pacific Northwest Research Institute. Clamcer and other things. Opinions my own.
Helen Hay Whitney Foundation fellow in Harmit Malik lab at Fred Hutch | I study evolutionary adaptation | http://tamanashbhattacharya.wordpress.com
Mol bio | Genomics | Development | AI
PhD @Radboud uni, postdoc @ EMBL, Genomics scientist @Google DeepMind
Fragile Nucleosome co-organiser
Views are my own
Animal lover, PNWer (currently Bay Area), structural biologist @ Genentech
🧪🏃🏻♀️
MCB PhD student @ UW/Fred Hutch
🔬 Assistant Prof, Pathology @Duke | Director, Clin Micro Lab
🧫 Former Clin Micro Fellow @Memorial Sloan Kettering
👩🏻🔬 Former Postdoc @broadinstitute.org
🎓 PhD @The Rockefeller University
Focus: Diagnostics, AMR, Structural Biology
EPIGENETIC HULK SMASH PUNY GENOME. MAKE GENOME GO. LOCATION: NOT CENTROMERE, THAT FOR SURE
Stanford BioE, Genetics & Sarafan ChEM-H. Chan-Zuckerberg Biohub Investigator. Our lab develops and applies microfluidic assays for high-throughput biophysics and biochemistry.
Associate Professor / enhancers - 3D genome - morphogenesis
Website: https://www.unige.ch/medecine/gede/en/research-groups/999andrey
Incoming Associate Professor @ Colorado University Anschutz | cancer epigenetics | DNA replication | protein synthesis | vanrechemlab.com
A bit of 3D gene regulation, single-cell omics and transgenic models.
Born and raised in the bay of Algeciras. Enjoying Science and Flamenco at CABD, Seville.
Lab website: https://lupianezlab.github.io/Website/
Email: dario.lupianez@csic.es
Assistant Professor, UBC school of Biomedical Engineering. Trying to enable personalized medicine by solving gene regulatory code.
Scientist in the School of Biotechnology and Biomolecular Sciences (BABS) at UNSW Sydney studying gene regulation and transcription factors in adipose tissue-resident immune cells and red blood cells
Deputy Director at WEHI, Melbourne Australia.
Lab head studying epigenetic control, in the context of X inactivation, genomic imprinting, SMCHD1, Prader Willi Syndrome, FSHD.
Mum, wife, beach lover.
Lecturer, researcher in CRISPR gene editing, epigenetics etc, and university administrator