
New Practical Cheminformatics Post
patwalters.github.io/Three-Papers...
@wpwalters.bsky.social
Cheminformatics, ML, Drug Discovery

New Practical Cheminformatics Post
patwalters.github.io/Three-Papers...

New Practical Cheminformatics Post, "Useful RDKit Utils - A Mötley Collection of Helpful Routines"
patwalters.github.io/Useful-RDKit...
The latest Practical Cheminformatics post, “The Trouble With Tautomers,” emerged from a discussion about the impact of tautomers on machine learning model predictions.
patwalters.github.io/The-Trouble-...
Thanks, Colab can be picky
24.03.2025 14:59 — 👍 0 🔁 0 💬 0 📌 0
The latest addition (#33) to the Practical Cheminformatics Tutorials series explores Bayesian optimization of reaction conditions.
github.com/PatWalters/p...
You know how I feel about bar plots for showing distributions, right? 😀
12.03.2025 00:16 — 👍 0 🔁 0 💬 1 📌 0
In a new Practical Cheminformatics post titled "Even More Thoughts on ML Method Comparisons," I share several plots that I find valuable for comparing machine learning methods.
practicalcheminformatics.blogspot.com/2025/03/even...
Determine bond order from XYZ format of molecule #RDKit
As readers know that recent version of RDKit supports XYZ format which is used in quantum chemistry field. The format doesn't have information of bond order. So to re-construct from the information bond order determination step is required.…
All of it! Lots of great new datasets. More new blogs coming online with great content. Open science at its best!
25.01.2025 16:25 — 👍 1 🔁 0 💬 1 📌 0
Machine Learning in Drug Discovery Resources page updated for 2025. github.com/PatWalters/r...
23.01.2025 14:12 — 👍 94 🔁 28 💬 5 📌 3
Deep learning for proteins tutorial: github.com/Graylab/DL4P...
28.12.2024 03:14 — 👍 67 🔁 28 💬 1 📌 0Super cool!
15.12.2024 13:06 — 👍 1 🔁 0 💬 1 📌 0
ChEMBL 35 is out. Happy Holidays!
chembl.blogspot.com/2024/12/here...
Thank for the invitation @freundlichgroup.bsky.social. I had a blast!
12.12.2024 03:37 — 👍 1 🔁 0 💬 0 📌 0Yes, absolutely! The approach was designed to facilitate interpretation by med chemists.
11.12.2024 03:37 — 👍 1 🔁 0 💬 0 📌 0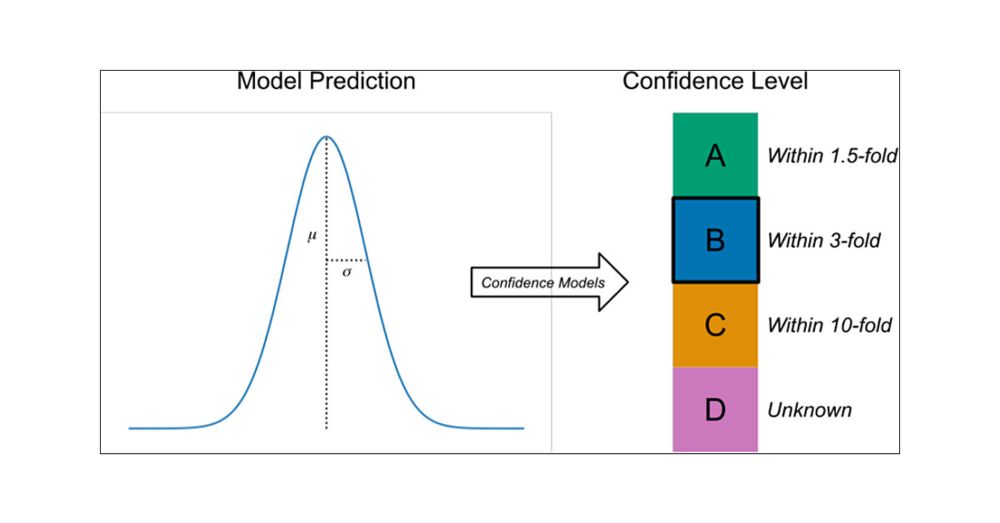
Excellent new paper (with code) by my former colleagues Steven Kearnes and Patrick Riley describing a procedure for associating confidence levels with regression model predictions in drug discovery. pubs.acs.org/doi/10.1021/...
10.12.2024 13:02 — 👍 26 🔁 11 💬 1 📌 0Well said, thank you Rommie!
09.12.2024 03:48 — 👍 7 🔁 0 💬 0 📌 0I'm not happy with the way this all went down. I have tremendous respect for Gabriele and the rest of the DiffDock authors. Their work has broken new ground and helped advance machine learning in drug discovery. If I had to do it over again, I'd do things differently.
09.12.2024 01:17 — 👍 18 🔁 2 💬 1 📌 1@prof-ajay-jain.bsky.social's response is here. www.linkedin.com/pulse/more-r...
09.12.2024 01:17 — 👍 5 🔁 2 💬 1 📌 1My "response to the response" is here. www.linkedin.com/pulse/my-res...
09.12.2024 01:17 — 👍 9 🔁 2 💬 1 📌 1Many of you have seen Gabriele Corso's response to our recent preprint. If you haven't, it's here. www.linkedin.com/pulse/respon...
09.12.2024 01:17 — 👍 16 🔁 1 💬 2 📌 0 08.12.2024 00:35 — 👍 2 🔁 1 💬 0 📌 0
08.12.2024 00:35 — 👍 2 🔁 1 💬 0 📌 0
I don't know. All the DiffDock results we report are from the original DiffDock paper.
06.12.2024 23:52 — 👍 3 🔁 0 💬 0 📌 0This is great, thanks for sharing
06.12.2024 10:58 — 👍 0 🔁 0 💬 0 📌 0Point taken
05.12.2024 16:43 — 👍 1 🔁 0 💬 0 📌 0For AutoDock-Vina, we directly compared with what was in the DiffDock paper.
05.12.2024 16:42 — 👍 1 🔁 0 💬 1 📌 0
I'm thrilled to announce a new preprint describing collaborative work with Ajay Jain and Ann Cleves Jain, "Deep-Learning Based Docking Methods: Fair Comparisons to Conventional Docking Workflows".
arxiv.org/abs/2412.02889
These have different uses. The one liner gets the SMILES for a particular ligand. My script uses that SMILES to assign bond orders, which are missing in PDB files.
02.12.2024 04:16 — 👍 1 🔁 0 💬 0 📌 0I prefer to think of myself as a realistic enthusiast 😆
22.11.2024 19:55 — 👍 14 🔁 0 💬 1 📌 0Hey Polaris!
21.11.2024 21:16 — 👍 1 🔁 0 💬 0 📌 0