Honored to serve @genomebiolevol.bsky.social and the greater @official-smbe.bsky.social community! 🫡
08.02.2026 00:02 — 👍 12 🔁 5 💬 1 📌 0

Homepage - Yeast Genetics Meeting 2026
Visit our website to learn more.
Honored to serve on the Yeast Genetics Meeting Program Committee of Evolution & Population Genetics. Welcome for meeting registration and abstract submission, especially on our Evolution & Population Genetics program! genetics-gsa.org/yeast-2026/
@genetics-gsa.bsky.social #Yeast26
30.01.2026 06:14 — 👍 5 🔁 2 💬 0 📌 0


First #SMBE meeting for our lab!Definitely a memorable experience. Already looking forward to the next one! @official-smbe.bsky.social #SMBE2025
25.07.2025 15:47 — 👍 8 🔁 2 💬 0 📌 0
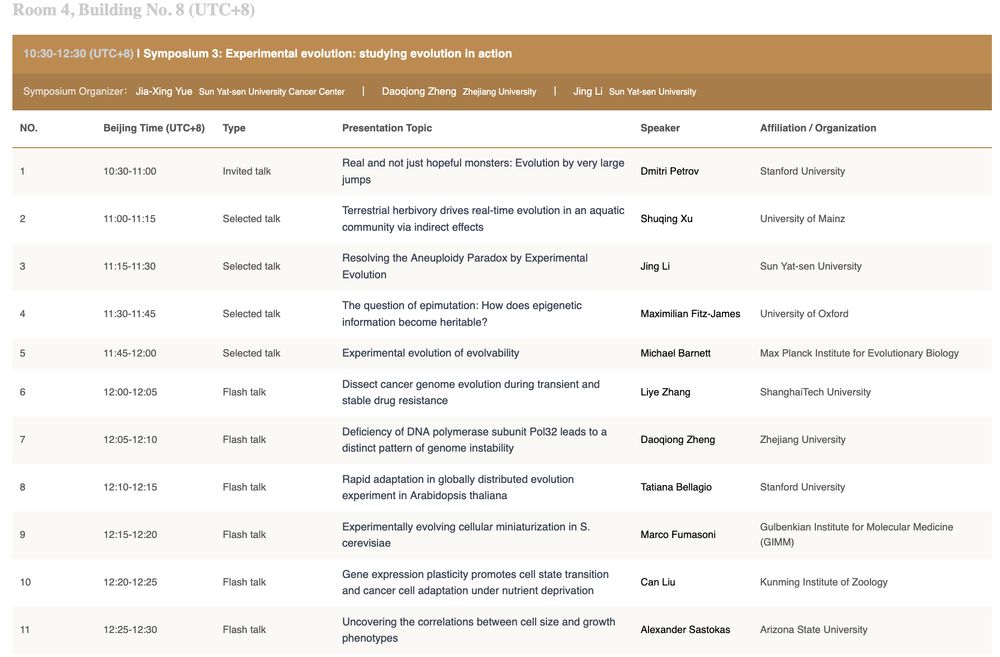
📣Our #SMBE 2025 Symposium (No. 3) on Experimental Evolution will be held today (7.22, Tuesday) at 10:30 in Room 4, Building No. 8. An associated poster session will be held at 14:00 (poster number: S3-P01~P16). Welcome to join! let's study evolution in action! 🥳 🎉 @official-smbe.bsky.social
21.07.2025 23:20 — 👍 7 🔁 4 💬 0 📌 0
Welcome to attend our SMBE2025 Symposium on Experimental Evolution. Co-organized by Dao-Qiong Zheng, @jingli-0512.bsky.social and myself.
17.06.2025 01:45 — 👍 6 🔁 2 💬 0 📌 0
Evomics Lab – Exploring Omics in the Light of Evolution
📢 3 Postdoc positions available in @evomicslab.bsky.social
targeting on genome instability evolution of both yeast and human genomes. Both computational genomics and experimental genomics backgrounds are welcome! 🧬🧑💻🧪 For more information, see evomicslab.org or send me a message!
28.04.2025 06:17 — 👍 5 🔁 5 💬 0 📌 0
📢📢📢 The FINAL COUNT DOWN for the extended abstract submission deadline of #SMBE2025! Please submit your abstract by 23:59 (GMT +8) of February 15, 2025 [Saturday]! And shout out again for our symposium on Experimental Evolution ! 🎉🎉🎉 @OfficialSMBE
#LTEE #ExperimentalEvolution
12.02.2025 12:58 — 👍 3 🔁 2 💬 0 📌 0
ScRAPdb
Finally, we would like to thank @HumanPangenome
for making the #HPRC resources available, which helped us a lot in testing and perfecting our VRPG. And of course, our own ScRAP #yeast pangenome collection reported at evomicslab.org/db/ScRAPdb/ and www.nature.com/articles/s41...
14.01.2025 05:59 — 👍 0 🔁 0 💬 0 📌 0
Many thanks to our talented student Zepu Miao for leading the development of VRPG. Built upon VRPG, we are currently working on new development and discoveries with #pangenome graphs. Stay tuned!
14.01.2025 05:59 — 👍 0 🔁 0 💬 1 📌 0
VRPG
To further demonstrate its functionality & scalability, we applied VRPG to the cutting-edge yeast and human reference pangenome graphs via a dedicated web portal at evomicslab.org/app/vrpg/
14.01.2025 05:59 — 👍 0 🔁 0 💬 1 📌 0
Especially, VRPG enables side-by-side visualization between the graph-based pangenome representation and the conventional primary-linear-reference-genome-based feature annotations, which greatly increases the Interpretability of pangenome graphs.
14.01.2025 05:59 — 👍 0 🔁 0 💬 1 📌 0
VRPG provides efficient & intuitive supports for exploring and annotating pangenome graphs along a linear-genome-based coordinate system and offers many highly useful features such as in-graph path highlighting, CNV and SV characterization, graph-based sequence query, etc.
14.01.2025 05:59 — 👍 0 🔁 0 💬 1 📌 0
To facilitate better exploration & understanding of pangenome graphs towards novel biological insights, here we present a web-based interactive Visualization and interpretation framework for linear-Reference-projected Pangenome Graphs (VRPG).
14.01.2025 05:59 — 👍 0 🔁 0 💬 1 📌 0
In the era of #T2T genomics, #pangenome graphs have emerged as a new paradigm for identifying, encoding, and presenting genomic variation at population levels. However, it remains challenging to dissect and interpret pangenome graphs in a biologically informative manner.
14.01.2025 05:59 — 👍 1 🔁 0 💬 1 📌 0
Welcome to join our dedicated symposium on Experimental Evolution for the @MolBioEvol 2025 Beijing Meeting! See you in Beijing!
13.12.2024 07:53 — 👍 0 🔁 0 💬 0 📌 0
As real case demonstrations, we applied NanoTrans to the public DRS datasets of yeast, Arabidopsis, as well as human embryonic kidney and cancer cell lines to showcase its utility, effectiveness, and efficacy across diverse systems and application settings.
10.12.2024 03:19 — 👍 0 🔁 0 💬 1 📌 0
Starting from the raw sequencing reads, NanoTrans can perform read basecalling, isoform clustering & quantification, differential gene expression & splicing, RNA modification, poly(A) tail length profiling, and fusion gene detection in a highly streamlined fashion.
10.12.2024 03:19 — 👍 0 🔁 0 💬 1 📌 0

ScRAPdb: an integrated pan-omics database for the Saccharomyces cerevisiae reference assembly panel
Abstract. As a unicellular eukaryote, the budding yeast Saccharomyces cerevisiae strikes a unique balance between biological complexity and experimental tr
Excited to introduce the ScRAPdb work that we recently published at Nucleic Acids Research, in which we assembled a #pangenome x #multi-omics database for #yeast. This is one step forward for a pan-multi-omics understanding of genome & trait evolution. Read more > academic.oup.com/nar/advance-...
17.11.2024 02:56 — 👍 5 🔁 4 💬 0 📌 0
Evolutionary microbiologist. Bacterial social life, virulence, drug resistance.
Assistant professor of Mycology and Plant Pathology, University of Tehran
🌱🍄💫
👩🏼💻 Science communication & media relations @embo.org | 🎓 Senior Associate @lucycavcoll.bsky.social @cam.ac.uk | 🦠 PhD in Molecular Virology |📍Heidelberg, Germany | Views my own
https://www.linkedin.com/in/astrid-gall-ab4787138/
EMBO is the organization of more than 2,100 leading researchers that promotes excellence in life sciences in Europe and beyond.
https://www.embo.org/
Evolutionary epigenomics ( eukaryotes / Transcription Factors / Transposable Elements / DNA methylation ) @ QMUL (London).
Lab website: https://www.demendozalab.com/
We are a lab focused on the study of invasive soil invertebrates, mainly land planarians. Led by Marta Álvarez Presas and based at the Universitat de Barcelona
Principal Investigator, Institute of Zoology, Chinese Academy of Sciences.
Transposable element and host-parasite interaction
Lab website: http://english.ioz.cas.cn/sourcedb/scs/202310/t20231030_431161.html
Prof at Cornell, #firstgen, immigrant 🇫🇷🇺🇸. Transposons, viruses, and all the cool stuff genomes are made of. https://www.feschottelab.com
🧬 Professor of Evolutionary Genomics at @zoology.ubc.ca and @ubcbiodiversity.bsky.social. Editor-in-Chief of @evolletters.bsky.social. Science with a sprinkling of dachshund pictures. https://www.zoology.ubc.ca/mank-lab/
ICREA Evolutionary Biologist working on transcriptomics of development and evolution at @upf.edu and @crg.eu. Coordinator of the @evomg-bcn.bsky.social Joint Program.
Lab website: http://transdevolab.com
Engineer and molecular biologist. Curious about the world and optimistic to make it better.
Evolutionary biologist at Southwest University
Genomics, big data, open science, diversity. Director of the Centre for Population Genomics, focused on building a more equitable future for genomic medicine. Opinions my own.
Population and evolutionary genetics @UCDavis. Posts, grammar, & spelling are my views only. He/him. #OA popgen book https://github.com/cooplab/popgen-notes/releases
lover of birds, nature, evolutionary biology, diversity in STEM, peace and good will
Bren Professor of Computational Biology @Caltech.edu. Blog at http://liorpachter.wordpress.com. Posts represent my views, not my employer's. #methodsmatter
Professor at Zhejiang University; sex chromosomes and sex determination; qizhoulab.org