
Our Science paper is out! 😃
www.science.org/doi/10.1126/...

Our Science paper is out! 😃
www.science.org/doi/10.1126/...

Gisela Otten has joined the Board of Directors and will Chair the Board of the Women’s team at Cambridge United Football Club. Gisela joined @felixrandow.bsky.social's group at the LMB in 2017 and spent 6 years playing for Cambridge United Women. #LMBAlumni
www.cambridgeunited.com/news/mike-da...

Another insightful piece recently out @natsmb.nature.com delineating how Shigella utilises its effector IpaH1.4 to counteract host RNF213-induced LPS ubiquitylation. Very interesting work by the lab of @felixrandow.bsky.social. If you want to, have a go at it: www.nature.com/articles/s41...
15.04.2025 09:03 — 👍 8 🔁 2 💬 0 📌 0
Comparison of Salmonella and Shigella host cell invasion. Salmonella are targeted by the host E3 ligase RNF213, which ubiquitylates bacterial lipopolysaccharide (LPS), marking bacteria for autophagic destruction. Shigella employs its effector proteins IpaH1.4 - an E3 ubiquitin ligase itself - to ubiquitylate RNF213, leading to its proteasomal degradation and thus preventing the host from tagging Shigella for destruction.

Cryo-EM structure showing how the leucine-rich repeat (LRR) domain of IpaH1.4 binds to the RING domain of RNF213
How do cytosol-invading bacteria evade LPS ubiquitylation by the host's defence machinery?
@felixrandow.bsky.social's group determined that Shigella flexneri uses IpaH1.4 to degrade the LPS ligase RNF213, inhibiting the cell's ubiquitylation abilities.
Read more: tinyurl.com/3y4kv2bp
#LMBResearch🧪

Don't miss similar findings from the Coers and Pan labs in BiorXiv and NatComm. Also note that Burkholderia antagonizes LPS ubiquitylation through DUB action (Thurston lab) and that Chlamydia deploys GarD (Coers lab). Conclusion: Bacteria decided RNF213 is a hot target in host–pathogen warfare!
09.04.2025 21:44 — 👍 6 🔁 0 💬 0 📌 0
Remarkably, IpaH1.4 doesn’t stop at RNF213—it targets several host E3 ligases via their RING domains. Individual RINGs bind overlapping sites on IpaH1.4 through unique interactions, explaining specificity of IpaH1.4 towards select RING domains. A clever strategy!
09.04.2025 21:44 — 👍 5 🔁 0 💬 2 📌 0
Cryo-EM reveals IpaH1.4 binds the RING domain of RNF213—a region with no known E3 activity in RNF213. However, despite pressure from IpaH1.4, the RING is highly conserved, hinting at a key function. Intriguingly, Moyamoya disease–linked mutations also occur in this domain.
09.04.2025 21:44 — 👍 3 🔁 0 💬 1 📌 0
RNF213, a host E3 ligase, ubiquitylates LPS to mark cytosol-invading bacteria for autophagy. But how do cytosol-adapted bacteria escape? We found that Shigella's E3 ligase IpaH1.4 blocks LPS ubiquitylation by degrading RNF213 via the proteasome.
www.nature.com/articles/s41...

Not long until the deadline for this faculty position @dunnschool.bsky.social...
Are you the one to lead a group doing great research in a special environment, while teaching outstanding students?
www.path.ox.ac.uk/vacancy/asso...

Exciting news: Registration for the 2025 UK Proteotasis & Autophagy meeting (3-5th June) is open: tinyurl.com/mr3p49f9
18.02.2025 09:32 — 👍 5 🔁 3 💬 0 📌 0

There are few moments more rewarding in running a lab than having a student defend their PhD thesis. Congratulations Dr. @marianapdc.bsky.social for this amazing achievement. And thanks to @lloydlab.bsky.social and Bart Lambrecht for being the examiners.
07.02.2025 20:45 — 👍 27 🔁 5 💬 2 📌 2
Lara Krüger, stood in the LMB atrium, smiling.
Congratulations to @lara-kruger.bsky.social, postdoc in @deriverylab.bsky.social in @cellbiol-mrclmb.bsky.social, who is leaving the LMB to establish her own research group at @institutcurie.bsky.social in Paris!
Best of luck to you Lara!
#LMBAlumni
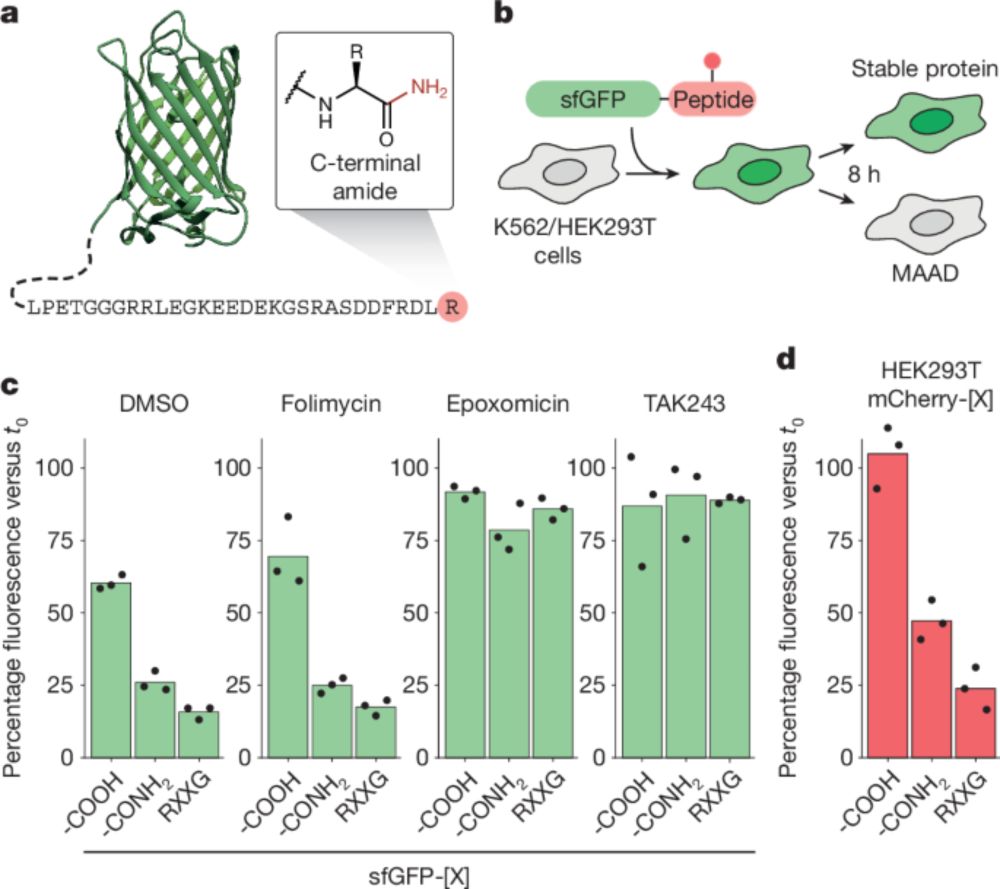
How do cells recognise disease- and stress-affected proteins for clearance by ubiquitination? Work @nature shows that such proteins are marked with C-terminal amides, allowing their targeting by SCF/FBXO31 ubiquitin ligase for degradation. shorturl.at/B3rmR
& NV
shorturl.at/fImjA
Guess what? You got it, another @plosbiology.org focus issue of thought-provoking Perspectives and Essays on an issue relevant to this Keystone Symposium
What can I say? We clearly care a lot about many of the topics at these joint meetings
Take a look and let us know what you think! 👀⬇️
🧪
We made it to 100! 📢💯 The #ubiquitin and #ubl Starter Pack now includes 100 profiles to follow if you’re interested in any aspect of the field: signalling, structure and biochemistry, biology, mechanisms, proteostasis, TPD, chemical biology… Check out the list and let me know if you are missing.
21.01.2025 11:10 — 👍 32 🔁 13 💬 1 📌 1Who’ll get the second popcorn??
17.01.2025 13:08 — 👍 1 🔁 0 💬 1 📌 0
By my reckoning, yesterday marked the 50th anniversary of ubiquitin's discovery! 🎈🎉🎁🥂
www.pnas.org/doi/10.1073/...

We just can't stop recruiting!
Associate professor in cell and molecular biology @dunnschool, with a preference for immunology, inflammation and/or infection - all defined broadly
Come and be our colleague
Deadline 28 Feb, please spread the word
www.path.ox.ac.uk/vacancy/asso...


After lots of teasing by @npariente.bsky.social I updated my profile picture. No more impersonation of my younger self!
12.01.2025 12:36 — 👍 6 🔁 0 💬 0 📌 0
PhDs in biochemistry, structural biology, pharmacology, or similar, located in the US and with strong publication track records and a high motivation to solve Parkinson's, please send CV directly to me at jimhurley@berkeley.edu.
Repost appreciated.
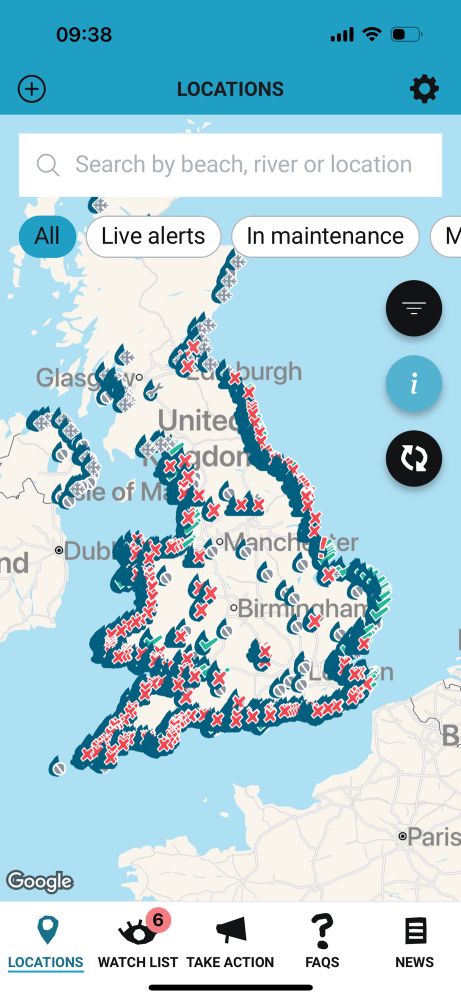
Real-time map from Surfers Against Sewage app. For those in blissful ignorance, this is what happens after normal heavy rains in Britain: huge swathes of UK coastline rendered unswimmable by water companies’ discharges. I know it’s old news but I refuse to become desensitized to the sheer awfulness
07.01.2025 09:50 — 👍 13 🔁 7 💬 1 📌 0
We found that many bacterial species use exogenous peptidoglycan fragments - released by lysis of neighboring cells - as a general danger signal, triggering a danger response that protects bacteria against many dangers: biofilm formation.
Details here 👇
www.nature.com/articles/s41...

As we go through various steps of the peer-review process, @gmivienna.bsky.social comms team put together a nice summary of our recent #preprints : www.oeaw.ac.at/gmi/detail/n... Thx @philippdexheimer.bsky.social for the illustration
07.01.2025 07:19 — 👍 37 🔁 12 💬 2 📌 0On Jan 6 1912, Alfred Wegener presented his theory of continental drift and supercontinent Pangaea's breakup into today's continents. Despite initial rejection, modern seismology and ocean drilling in the 1950s/60s validated his ideas, leading to the current understanding of plate tectonics. ⚒🧪
06.01.2025 09:19 — 👍 754 🔁 100 💬 23 📌 8Happy New Year! Let’s start with an updated starter pack. Now with 94 profiles to follow if you are interested in anything #ubiquitin or #Ubl: signalling, structure and biochemistry, biology, mechanisms, proteostasis, TPD, chemical biology, etc. Let me know if you or anyone else is missing.
06.01.2025 13:02 — 👍 13 🔁 7 💬 3 📌 0
Peptidoglycan as danger signal promotes biofilm formation - great article by the Drescher lab
www.nature.com/articles/s41...

New paper out from Deretic’s lab!!! rupress.org/jcb/article/...
02.01.2025 20:06 — 👍 5 🔁 2 💬 0 📌 0The pleasure was mine!
01.01.2025 21:21 — 👍 1 🔁 0 💬 0 📌 0
Today in A Coruña. It has become a nice tradition for us to visit their annual photo exhibition.
26.12.2024 18:20 — 👍 4 🔁 0 💬 2 📌 0Believe it or not, GRC has told me we need more registered applicants or this meeting might not survive! Please don't let that happen if you can afford to attend 🙏
19.11.2024 22:46 — 👍 25 🔁 29 💬 2 📌 2