GitHub - THGLab/HiQBind: Workflow to clean up and fix structural problems in protein-ligand binding datasets
Workflow to clean up and fix structural problems in protein-ligand binding datasets - THGLab/HiQBind
Check it out, and feel free to drop your questions here or on GitHub!
🔗 GitHub: github.com/THGLab/HiQBind
📄 Paper: pubs.rsc.org/en/content/a...
#machinelearning #proteinligand #proteinligandbindingaffinity #structuralbiology #ai4sci
07.04.2025 23:49 — 👍 0 🔁 0 💬 0 📌 0

GitHub - THGLab/HiQBind: Workflow to clean up and fix structural problems in protein-ligand binding datasets
Workflow to clean up and fix structural problems in protein-ligand binding datasets - THGLab/HiQBind
Huge shoutout to the amazing team behind this:
👏 Lead author Yingze (Eric) Wang and @kunyangsun.bsky.social
👏 PI Prof. Teresa Head-Gordon
👏 Teammates Jie Li, Xingyi Guan, Oufan Zhang, Dorian Bagni
👏 Collaborators Dr. Heather A. Carlson and Prof. Yang Zhang
07.04.2025 23:49 — 👍 1 🔁 0 💬 1 📌 0

GitHub - THGLab/HiQBind: Workflow to clean up and fix structural problems in protein-ligand binding datasets
Workflow to clean up and fix structural problems in protein-ligand binding datasets - THGLab/HiQBind
What’s next?
We’re exploring:
🔁 Rotamer refinement
🤖 Binding label extraction with LLMs (maybe 👀)
🧠 Better data splits (possibly inspired by PLINDER) to support ML research!
07.04.2025 23:49 — 👍 0 🔁 0 💬 1 📌 0

GitHub - THGLab/HiQBind: Workflow to clean up and fix structural problems in protein-ligand binding datasets
Workflow to clean up and fix structural problems in protein-ligand binding datasets - THGLab/HiQBind
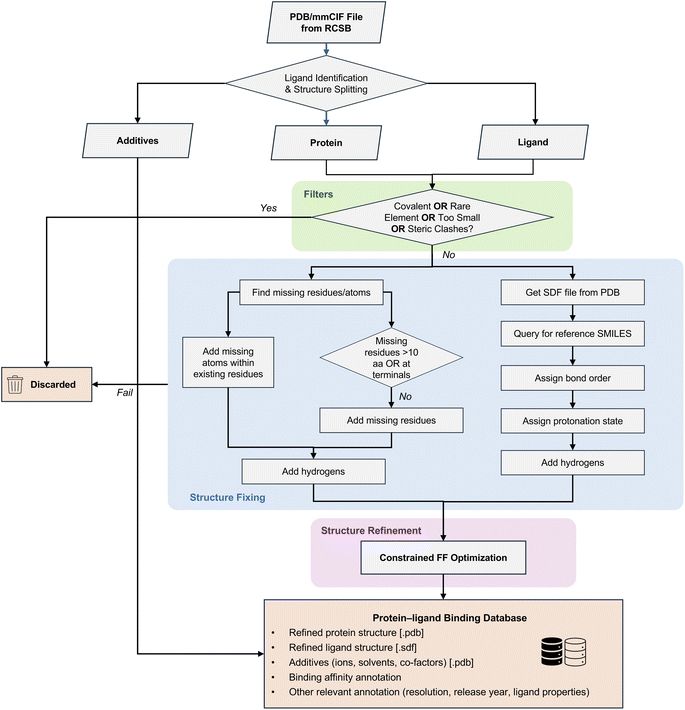
Since we're focused on structural data with binding labels, we applied this workflow to major open-access datasets (BioLiP, BindingDB, and BindingMOAD) to generate HiQBind: a cleaned, corrected dataset comparable in size to PDBBind v2020 but with significantly improved structural quality! 💥
07.04.2025 23:49 — 👍 1 🔁 0 💬 1 📌 0

GitHub - THGLab/HiQBind: Workflow to clean up and fix structural problems in protein-ligand binding datasets
Workflow to clean up and fix structural problems in protein-ligand binding datasets - THGLab/HiQBind
In this work, we built HiQBind-workflow, a semi-automated workflow that processes protein–ligand structures from the RCSB PDB by adding missing atoms, correcting ligand geometries, fixing bond orders and protonation states, and much more!
07.04.2025 23:49 — 👍 0 🔁 0 💬 1 📌 0
Copying Oliver's post from Linkedin to help us gain some visibility here!
🚨 Our paper is out! 🚨
"A workflow to create a high-quality protein–ligand binding dataset for training, validation, and prediction tasks" is now published in Digital Discovery! 🎉
07.04.2025 23:49 — 👍 2 🔁 1 💬 1 📌 0
We have left X for greener pastures and bluer skies - good riddance to Nazi-saluting Musk and his abuse of science and engineering that in fact enriched him.
20.03.2025 00:43 — 👍 7 🔁 0 💬 1 📌 0
Associate Professor @OIST, our group is interested in understanding the origin of protein function. mom, climber, skier
Flagship journal for @rsc.org, open access with no publication fee, all topics in chemistry. chemicalscience-rsc@rsc.org 🌐 Website: rsc.li/chemscience
Science writer and author of books including Bright Earth, The Music Instinct, Beyond Weird, How Life Works.
Ad Astra Fellow, Asst. Prof. of Digital Chemistry, @ucddublin.bsky.social |
Editor, @joss-openjournals.bsky.social |
Personal: ewcss.info |
Research group (@coreacter.org): CoReACTER.org |
ORCID: orcid.org/0000-0003-1554-197X |
All opinions mine
Associate Professor of Biochemistry at Virginia Tech. Comp Chem, CADD, and Drude FF contributor. Opinions my own. Go Hokies!
Computational chemistry and biophysics. Empirical force fields, CADD, RNA, carbohydrates, and methods developments including SILCS (site identification by ligand competitive saturation, See SilcsBio LLC) and enhanced-sampling GCMC. And I surf....
Quantum chemist at Cardiff
Tries to pass as a computational chemist. Bostero fanatic. He/Him. Comments are my own and do not reflect my institution.
Professor #compbiophys #compchem @UT_Dallas
QM/MM force fields DNA rep./modif., cancer biomarkers, ionic liquids, #SACNAS, views my own, he/él 🇲🇽🇺🇸
🇪🇸
Chemist • Professor at University of Vienna • Theoretical chemistry, quantum chemistry, excited-state dynamics •
Home of SHARC 🦈
Senior Editor of ACS Central Science •
Board of Directors, Cluster of Excellence MECS ☀️
She/her/wife/Mom of 2
Ab initio materials simulations at Duke University. Associate professor, Duke MEMS & Chemistry. https://aims.pratt.duke.edu/ . Opinions my own. Unlike the shirt on the right side of the profile image, which is not my own. The number on the shirt is 10.
Physicist, professor @univie.ac.at, director of @esivienna.bsky.social, computational physics, statistical mechanics, machine learning, soft matter, biking, hiking, skiing, *320 ppm. 🇪🇺
Professor, Georgia Tech Chemistry and Biochemistry. Georgia Research Alliance Eminent Scholar. Senior Editor, Protein Science, Editor-in-Chief, Electronic Structure. Pianist, photographer, foodie, dreamer, mom. She/they. #FirstGen #MENA All views my own.
Computational chemist at the University of Copenhagen #compchem
Research group lead by @bussigio.bsky.social at SISSA | #RNA, #moleculardynamics, and more | web: http://bussilab.org
Professor @pittchem.bsky.social. Computational biophysicist, leading @westpasoftware.bsky.social development for weighted ensemble rare-event sampling, Amber force field developer.
Computational Materials Science. Assistant Professor at UC Berkeley.
Assistant Professor at UC Berkeley
Research scientist & computational chemist at Berkeley Lab using HT DFT workflows, machine learning, and reaction networks to model complex reactivity.






