
Congratulations to Mike MacCoss on receiving the Donald F. Hunt Distinguished Contribution in Proteomics Award from US HUPO!
us-hupo.org/Distinguishe...
@chrismcgann.bsky.social
Postdoctoral Research Fellow, Harvard Medical School & Dana-Farber Cancer Institute

Congratulations to Mike MacCoss on receiving the Donald F. Hunt Distinguished Contribution in Proteomics Award from US HUPO!
us-hupo.org/Distinguishe...

Congratulations to Bill Noble on receiving the 2026 Gil Omenn Computational Proteomics Award from US HUPO!
us-hupo.org/Computationa...

If you’re at #HUPO2025, be sure to stop by my poster and learn about the latest developments in the BioPlex project!
12.11.2025 15:26 — 👍 9 🔁 3 💬 0 📌 0

More #gastruloid studies. Benchmarking correlations between RNA/proteins w/ acquiring information @ phosphorylation across #mouse & #human www.biorxiv.org/content/10.1...
Also looking at #Disease modelling. Fantastic #TourDForce Valuable bench marking and insight
@dschweppe.bsky.social #SCBEMs

Fantastic project led by @bo-wen.bsky.social. Excited to see the future uses of AI and transfer learning in proteomics. #massspec #proteomics
www.nature.com/articles/s41...

Excited to see this published in JPR. For years I've wanted a simple way to standardize the signal between instruments. We use the precision of an intraspectrum ratio to assess the relationship between the reported signal and the number of ions. pubs.acs.org/doi/10.1021/...
22.10.2025 15:32 — 👍 33 🔁 6 💬 1 📌 0
Can nanoparticles help us probe the glycoproteome?
The new pre-print from @riley-research.bsky.social seeks to answer this question using the Proteograph technology from Seer Inc.
Check out this lovely study on glycoproteins NP-enriched from biofluids here: chemrxiv.org/engage/chemr...

Real-time API control of mass specs has been hard to get started (C#, VS, etc). Mike Hoopmann's recent work makes dev easier & more open!:
Nova - lightweight dev library
schweppelab.github.io/Nova/ pubs.acs.org/doi/full/10....
Helios - a unified instrument API now @ JPR
github.com/SchweppeLab/...
Thanks William!
06.09.2025 00:48 — 👍 0 🔁 0 💬 0 📌 0Thanks!! I’m back to Boston for a postdoc!
06.09.2025 00:48 — 👍 1 🔁 0 💬 0 📌 0
After five great years, my journey in the Schweppe Lab and at UW has come to a close. Huge thanks to @dschweppe.bsky.social and all the amazing labmates for making graduate school such a wonderful experience. Extremely grateful for the memories and all the help I received along the way.
05.09.2025 15:56 — 👍 30 🔁 1 💬 3 📌 0
(JASMS) [ASAP] Dynamic Quadrupole Selection to Associate Precursor Masses with MS/MS Products in Data-Independent Acquisition: Journal of the American Society for Mass SpectrometryDOI: 10.1021/jasms.5c00110 (RSS) #MassSpecRSS #JASMS
09.08.2025 20:05 — 👍 4 🔁 1 💬 0 📌 0
News in Proteomics Research blog post | Nova! Build all the mass spec tools! proteomicsnews.blogs...
---
#proteomics #prot-other

"I invite all researchers to enter the glycoproteomics field and join the battle.”
With the complexity of glycoproteomics and how little we still know, Tim Veth puts out a rallying cry to speed up development in this promising field.
Learn more about Tim & read our discussion: bit.ly/3IXpo8B

Nova: A Library for Rapid Development of Mass Spectrometry Software Applications pubs.acs.org/doi/10....
---
#proteomics #prot-paper

Announcing the yEvo Mutation Browser, a Shiny app for visualizing and exploring sequencing data from experimental evolution and genetic screens. This is the latest from our @yevolab.bsky.social high school teaching & research project and led by my grad student Leah. www.biorxiv.org/content/10.1...
22.07.2025 12:53 — 👍 17 🔁 8 💬 2 📌 0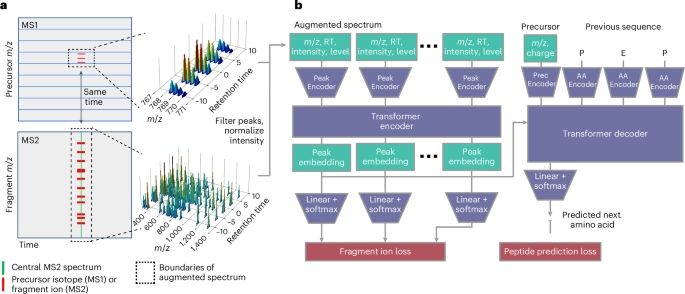
If you asked me 5 years ago if it would be possible to use a de novo tool on DIA data, I would have thought it would only exist in science fiction. Love being proved wrong. Great work from Justin Sanders. #proteomics #massspectrometry
www.nature.com/articles/s41...
Thanks so much @dschweppe.bsky.social!!! Couldn’t ask for a better place to do graduate school than the Schweppe Lab!
26.06.2025 15:52 — 👍 13 🔁 1 💬 0 📌 0


Huge congratulations to @chrismcgann.bsky.social who passed his PhD dissertation defense yesterday with flying colors!!
Dr. McGann was the lab’s first PhD student and now first graduate! So excited and proud of all of the things he’s accomplished!
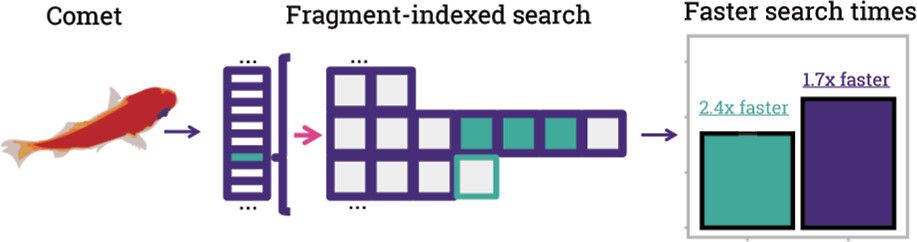
Comet's open-source Fragment-Ion Indexing work is live at JPR (@acs.org). @chrismcgann.bsky.social, Jimmy Eng and Erik Bergstrom showed how Comet could be dramatically sped up to keep pace with modern instrumentation and increasing sample sizes:
pubs.acs.org/doi/full/10....
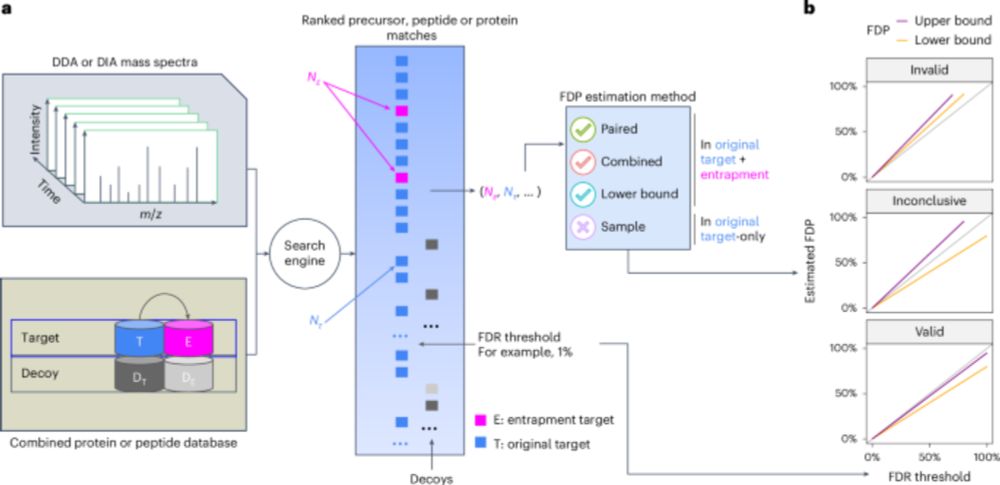
Assessing error control is fundamental in mass spectrometry-based proteomics. @bo-wen.bsky.social @maccoss.bsky.social @urikeich.bsky.social et al introduce a theoretical foundation for entrapment along with a method for more accurate evaluation of FDR control.
www.nature.com/articles/s41...

Congrats to @chrismcgann.bsky.social (and the other awardees) for receiving an @asms.org Graduate Student Travel Award!!
Photo credit to Katarina for capturing the moment!


Thanks to everyone who came out to the Real time MS bioinformatics hub yesterday! Great to chat about current and new directions! Here’s some action shots of Mathieu and Mike leading the discussion.
03.06.2025 15:28 — 👍 2 🔁 2 💬 0 📌 0
#ASMS2025 offers great evening workshops. If you are interested in real-time decision-making during MS data acquisition or still looking for one to join,
consider the Real time Mass Spectrometry Workshop featuring speakers and panelists Sarah Sipe, Aarthie Senathirajah, and Manuel Peris Diaz.

Nova: A library for rapid development of mass spectrometry software applications www.biorxiv.org/cont...
---
#proteomics #prot-preprint

A transformer model for de novo sequencing of data-independent acquisition mass spectrometry data
www.biorxiv.org/content/10.1...
impressive work!
03.03.2025 15:00 — 👍 2 🔁 0 💬 0 📌 0
This JAMA Insights explores the capability of proteomics to analyze thousands of proteins in patient samples, which could improve clinicians’ understanding of and ability to treat a wide range of diseases.
ja.ma/4jvNHbP
#MedSky
This prototype 908 devices SPE-CZE column and emitter combination looks to be the separation method to beat for localising phosphosites on multiply phosphorylated peptides.
13.12.2024 06:13 — 👍 13 🔁 2 💬 0 📌 0