
Reimagining Druggability using Chemoproteomic Platforms
Reimagining Druggability using Chemoproteomic Platforms - Innovative Genomics Institute (IGI)
I rarely get to give a research talk at my home institution! Looking forward to giving a talk about our latest research next Tuesday, November 18th, at 2 pm as part of the @innovativegenomics.bsky.social seminar series in Room 115 IGIB at UC Berkeley! innovativegenomics.org/events/semin...
11.11.2025 18:18 — 👍 6 🔁 1 💬 0 📌 0
Huge congrats to first author @cestieger.bsky.social for leading this effort!
Thanks to @novartis.bsky.social and @themarkfdn.bsky.social for their support.
#MolecularTherapeuticsInitiative #BerkeleyChemistry #BerkeleyMCB
22.10.2025 19:11 — 👍 1 🔁 0 💬 0 📌 0
This establishes a new therapeutic modality - using small-molecule-induced proximity for targeted epigenetic reprogramming - to silence transcriptional networks long considered out of reach.
A potential blueprint for pharmacologically “turning off” disease-driving transcription factors.
22.10.2025 19:11 — 👍 2 🔁 0 💬 1 📌 0
In androgen-independent prostate cancers expressing both AR and the “undruggable” truncation variant AR-V7, where AR antagonists inhibit only ~40% of AR activity...
Our AR/AR-V7 TRACER, which links an AR SARM to an MBD2 recruiter, suppresses more than 90% of AR transcriptional activity.
22.10.2025 19:11 — 👍 0 🔁 0 💬 1 📌 0
TRACERs enable epigenetic silencing of transcription factors through fully small-molecule control.
We demonstrate that TRACERs can potently and selectively repress the transcriptional programs of estrogen receptor (ER) and androgen receptor (AR/AR-V7).
22.10.2025 19:11 — 👍 1 🔁 0 💬 1 📌 0

Targeted Transcriptional Repression by Induced Proximity
Most cancer driving proteins remain undruggable due to the absence of ligandable pockets and their reliance on intrinsically disordered or protein-DNA/protein-protein interactions. Transcription facto...
We unveil TRACERs, Transcriptional Regulation via Active Control of Epigenetic Reprogramming: a new small-molecule-based induced-proximity modality that silences transcription factors by recruiting endogenous corepressor complexes for locus-specific repression.
www.biorxiv.org/content/10.1...
22.10.2025 19:11 — 👍 26 🔁 1 💬 1 📌 2
Haha I would never…
27.06.2025 12:41 — 👍 0 🔁 0 💬 1 📌 0
This paper now out in ACS Central Science!! pubs.acs.org/doi/10.1021/...
27.06.2025 05:35 — 👍 8 🔁 0 💬 0 📌 0
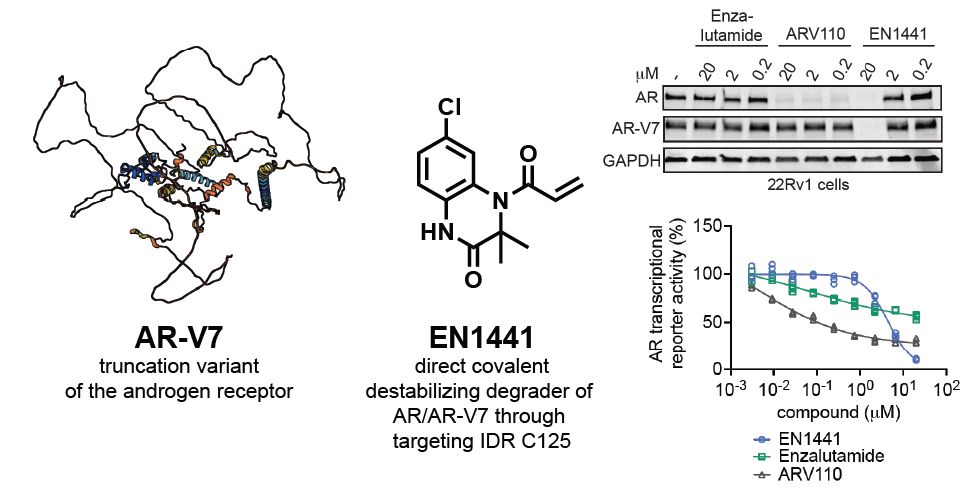
Our latest paper w/ @pottslab.bsky.social & @amgen.bsky.social in @jacs.acspublications.org by co-first authors @cmzammit.bsky.social & Cory Nadel on discovery of covalent destabilizing degrader of AR & AR-V7 in prostate cancer. Thanks also to @themarkfdn.bsky.social ! pubs.acs.org/doi/10.1021/...
10.06.2025 04:26 — 👍 18 🔁 5 💬 0 📌 0


Excited to announce a new collaboration between the Molecular Therapeutics Initiative (MTI) and Amgen Inc.! This partnership includes funding for cutting-edge research & an internship program for grad students to train in drug discovery & development at Amgen.
#MTI #Amgen #Biotech #UCB
08.06.2025 00:27 — 👍 6 🔁 1 💬 1 📌 1
Spirit
Preliminary score: ND
Bioorganic GRC is set! I got tired of the lack of swag so made a couple of unofficial tees. Order one if you want to support the conference (or simply enjoy strange looks in grocery stores).
Spirit: chembio-shop.fourthwall.com/products/grc...
World Tour: chembio-shop.fourthwall.com/products/spi...
03.06.2025 03:16 — 👍 41 🔁 6 💬 6 📌 4
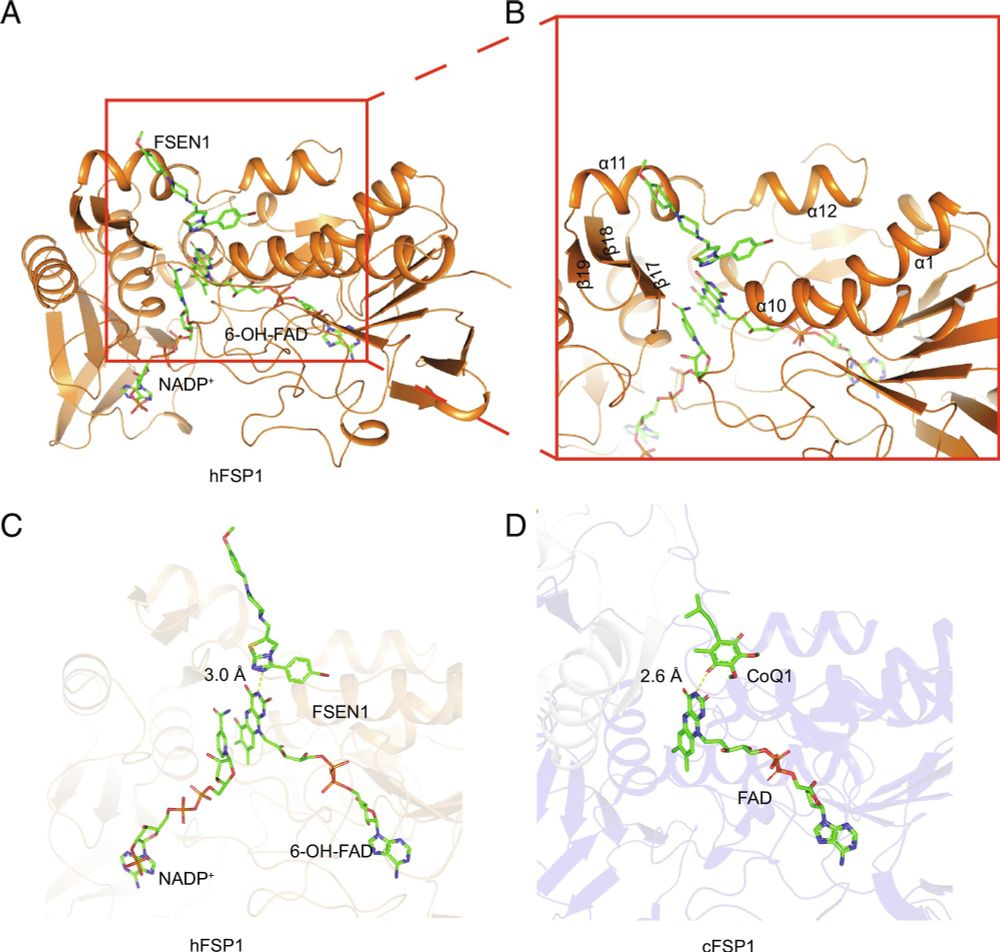
Cocrystal structure reveals the mechanism of FSP1 inhibition by FSEN1 | PNAS
FSP1 is an FAD-dependent oxidoreductase that uses NAD(P)H to regenerate the reduced
forms of lipophilic quinone antioxidants, such as coenzyme Q10 ...
Thrilled this paper is out! We solved the first cocrystal structure of FSP1 with an inhibitor (FSEN1), providing mechanistic insight & a foundation for medchem. Led by Amalia Megarioti & Sitao Zhang. A terrific collaboration with Da Jia.
Brief Skytorial!
1/9
29.05.2025 18:01 — 👍 76 🔁 29 💬 12 📌 2

Congrats to chem bio grad students Ellie King, Lauren Orr, and Vienna Thomas on graduating with their PhD's!
19.05.2025 20:49 — 👍 14 🔁 0 💬 0 📌 0

Our 2025 lab photo!
16.05.2025 02:42 — 👍 29 🔁 0 💬 0 📌 0
- Mutations mimicking glues by @brianliau.bsky.social al and Ning Zheng
- Covalent and bivalent glue engagement by @dannomura.bsky.social, Nathanael Gray, @georgwinter.bsky.social and @alessiociulli.bsky.social
- And finish it of with a discussion on general mode-of-action.
08.05.2025 09:27 — 👍 9 🔁 3 💬 1 📌 0
Molecular basis of SIFI activity in the integrated stress response - Nature
Nature - Molecular basis of SIFI activity in the integrated stress response
Excited that our work revealing the structure and function of the monster stress response silencing factor, the E3 ligase SIFI, is now out! Congratulations to the most amazing team of Zhi Yang, Diane Haakonsen, Michael Heider and Sam Witus!
www.nature.com/articles/s41...
07.05.2025 10:27 — 👍 45 🔁 14 💬 1 📌 2
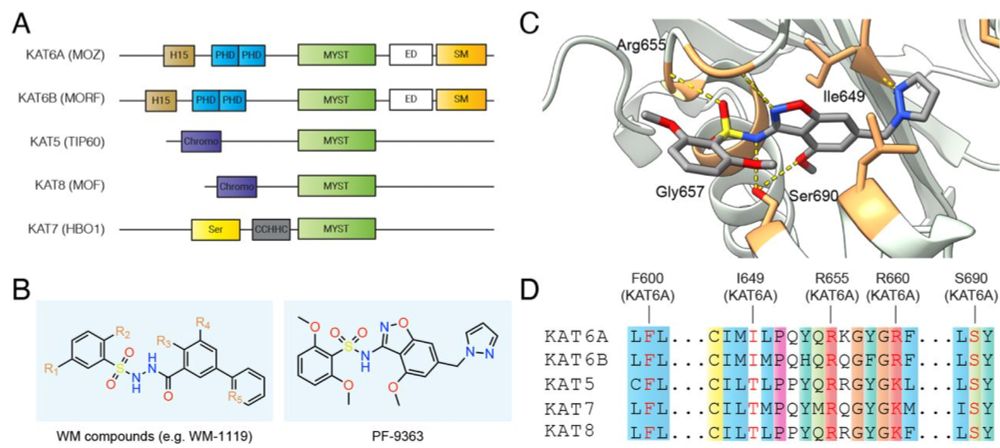
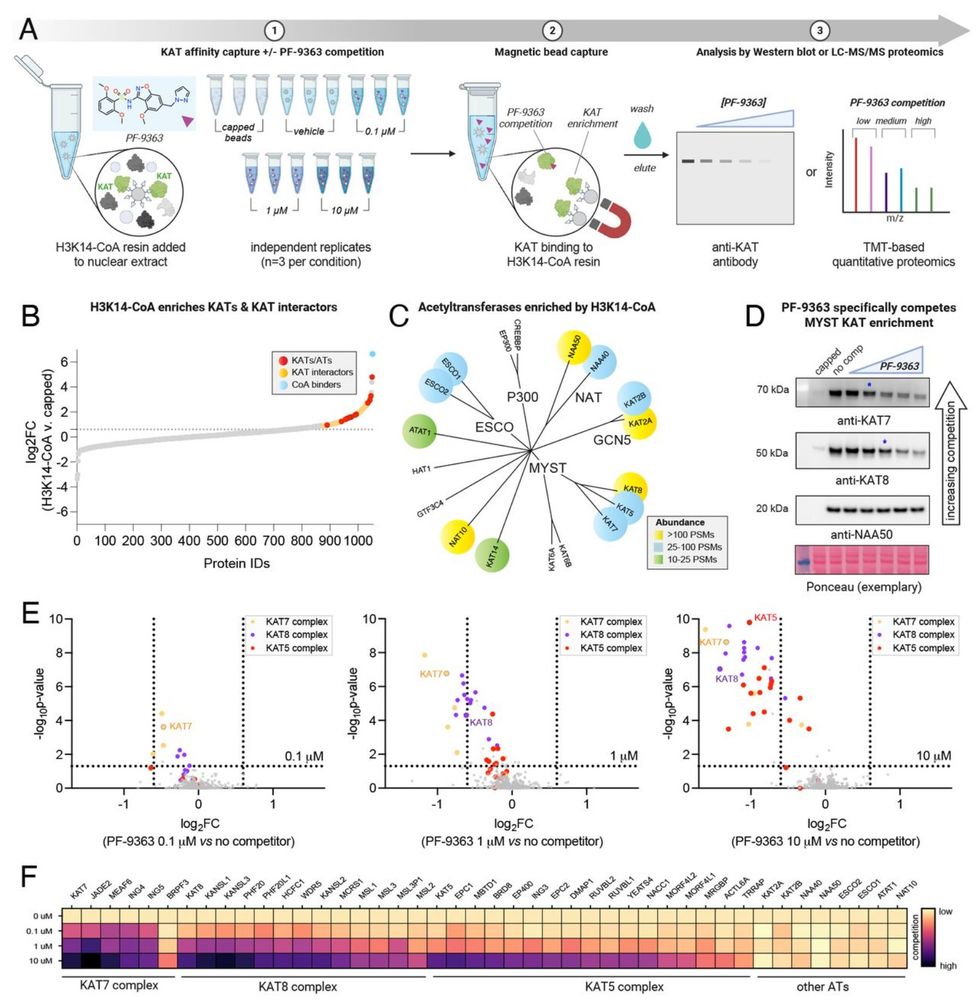
New-preprint alert! We use chemoproteomic profiling to show MYST KAT inhibitors exhibit hierarchical target engagement (KAT6A/B > KAT7 >> KAT8). Reminiscent of Type I kinase inhibitors, the selectivity of these small molecules varies based on concentration.
www.biorxiv.org/content/10.1...
04.05.2025 14:22 — 👍 30 🔁 4 💬 1 📌 0
Thanks to @olzmannlab.bsky.social for the great collaboration, and also thanks to @novartis.bsky.social @themarkfdn.bsky.social
26.04.2025 13:14 — 👍 2 🔁 0 💬 0 📌 0

On my way to #aacr25. I’ll be chairing Part 3 of “Chemistry to the Clinic” tomorrow (4/26 at 2:30) with talks from @dannomura.bsky.social and @xiaoyuzhang.bsky.social. Weather report calls for lots of chemoproteomics, molecular glues, and tough targets. Please join us!
26.04.2025 00:01 — 👍 19 🔁 2 💬 1 📌 0

Excited to moderate a panel at the @theaacr.bsky.social CICR townhall at #2025AACR on "Charting the Course: Innovating Cancer Drug Discovery in a Challenging Landscape" alongside Shiva Malek, Gwenn Hansen, Ryan Schoenfeld, and Joseph Pearlberg!
26.04.2025 02:29 — 👍 8 🔁 3 💬 1 📌 0

Back in Chicago for #2025AACR! I’ll be giving a talk on our latest work tomorrow in the Educational Session on Advances in Ligand Discovery to Enable Previously Intractable Targets on Sat Apr 26th 2:30-4:00pm in S406 McCormick South alongside @michael-erb.bsky.social & @xiaoyuzhang.bsky.social !
26.04.2025 02:23 — 👍 12 🔁 2 💬 0 📌 0


Amazing talk yesterday by @rushika-perera.bsky.social for our Molecular Therapeutics seminar series!
25.04.2025 18:01 — 👍 7 🔁 0 💬 0 📌 0
Editorial team of Nature Reviews Drug Discovery
www.nature.com/nrd
PhD student in the Crossley/Quinlan lab at UNSW Sydney. Interested in gene regulation 🧫, transcription factors, chromatin, and epigenetics. 🧬
Incoming assistant professor at Peking U | Postdoc in Kai Johnsson Lab at MPIMR Heidelberg | Ph.D in Xing Chen Lab at Peking U | Chemical biology, Expansion🔬, Molecular probes
scholar.google.co.kr/citations?user…
stanford md/phd student in the bintu lab, thinking about all things RNA in gene expression
Postdoc @UoD @CeTPD with Alessio Ciulli
Structural & chemical biology in Targeted Protein Degradation
Microbiologist. Infectious diseases. Drug discovery.
University of Tübingen, Germany
Official account for the Department of Chemistry at Stanford University, a world-class center for research and education in the chemical sciences.
Assistant Professor at Harvard Medical School
Chemical Biology, Cancer, Click Chemistry
Official Bluesky account of the Bendall Lab in the Stanford Department of Pathology and Immunology / Cancer Biology / Stem Cell research and training programs.
Prof. of Organic Chemistry⚗️ CATALYSIS & more (membranes, on-surface-chemistry, batteries, data & machine learning). Passionate 👨🔬😀🇪🇺
Assistant professor in Data Science and AI at Chalmers University of Technology | PI: AI lab for Molecular Engineering (AIME) | ailab.bio | rociomer.github.io
Student-run THG Lab account @UC Berkeley. We develop physics-based and machine learning-based models for various systems.
BIF PhD student in Schulman lab @mpibiochem.bsky.social, interested in structural biology and ubiquitin system
Stanford, cell cycle, RB, cancer, SCLC, mouse models
The Cress Lab | Innovative Genomics Institute @ UC Berkeley | Microbiome Editing | Microbiome Delivery Technologies | Phage and MGE Functional Genomics | Hiring!
https://www.cresslab.bio/
Postdoc in Jon Ostrem Lab at UCSF. Chemical biology, and targeted cancer therapies.
Chemical biologist #glycotime #chemsky #chembio
Big fan of total synthesis and catalysis | PhD from the Lautens group @UofT | Former undergraduate @UBC
Physician scientist studying ovarian cancer
https://obgyn.uchicago.edu/research/lengyelkenny-ovarian-cancer-laboratory
Group Leader, Induced Proximity Therapeutics, Centre for Protein Degradation, Institute of Cancer Research, London
























