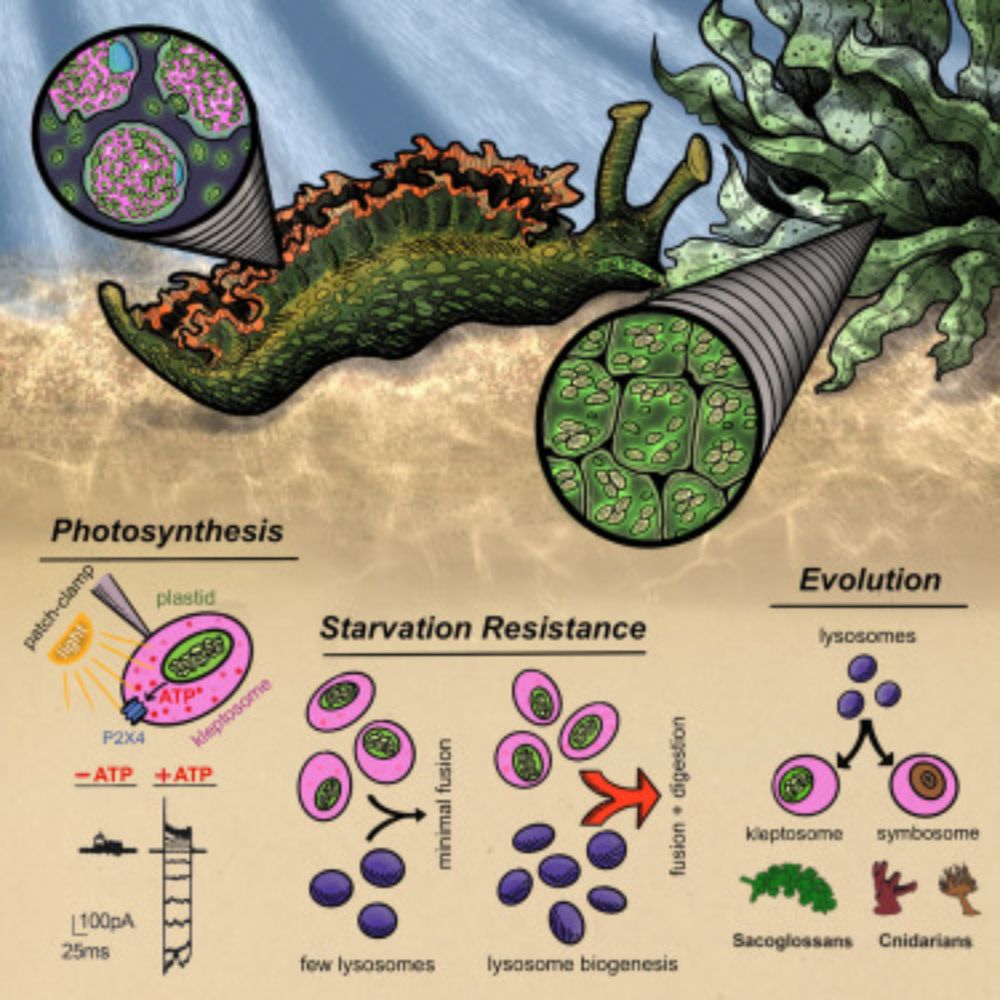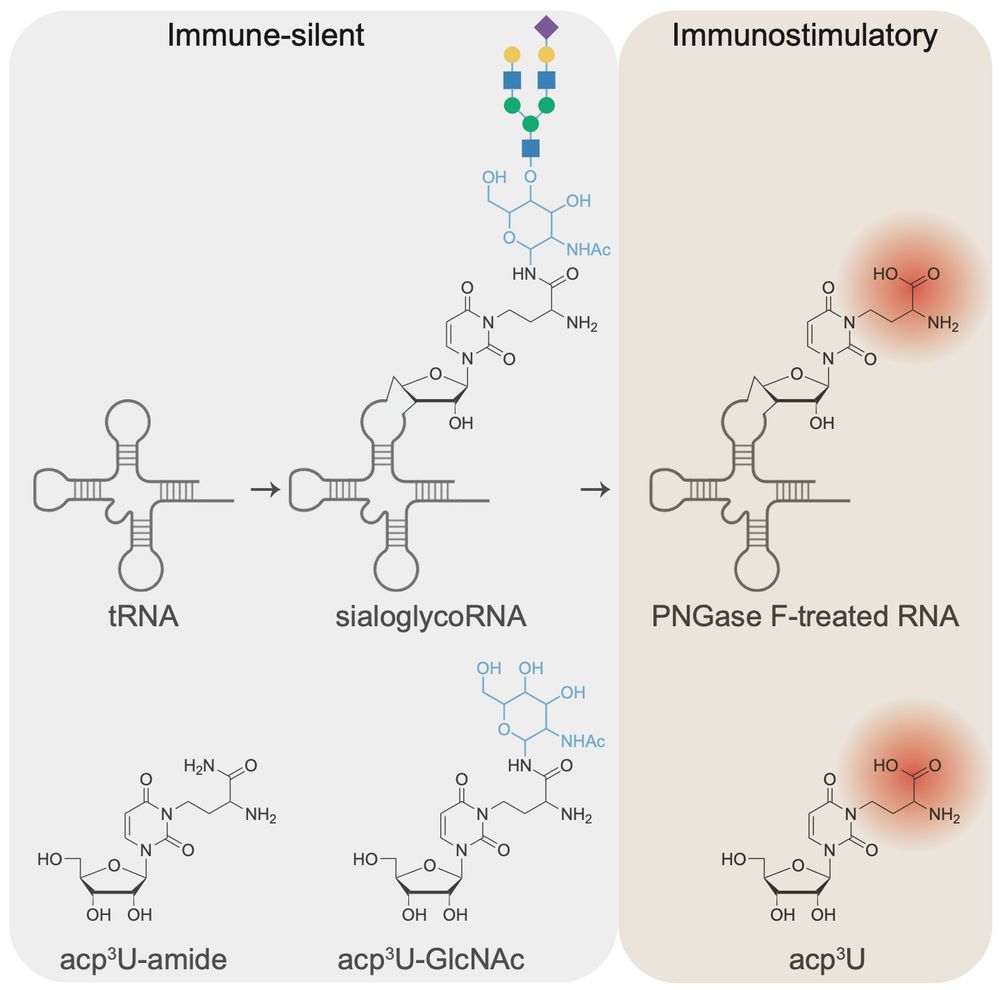
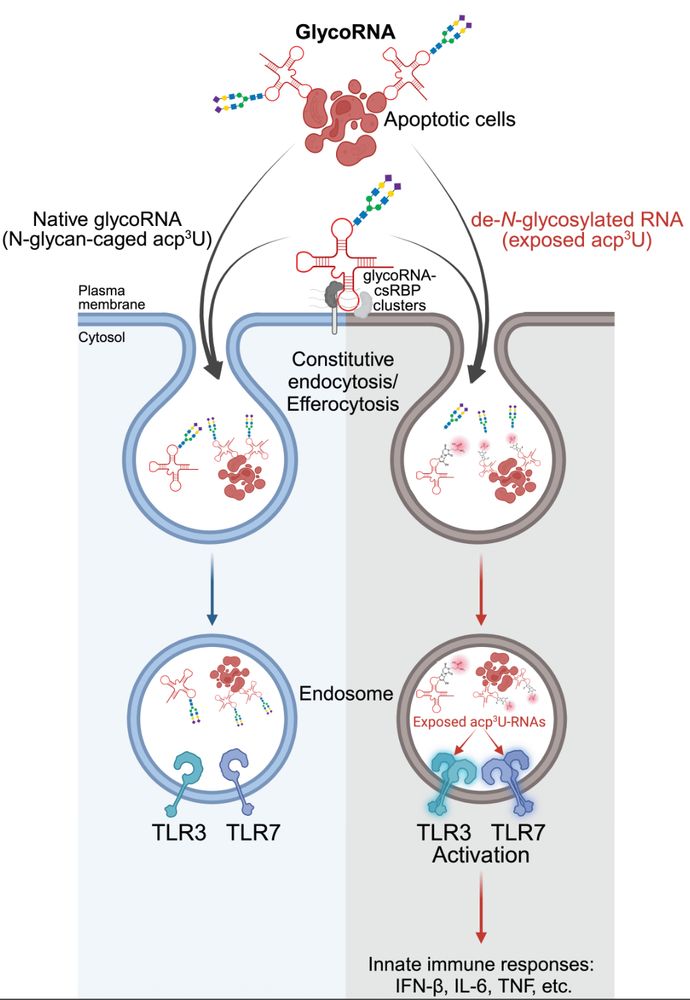
RNA N-glycosylation enables immune evasion and homeostatic efferocytosis by chemically caging acp3U. Excited to report this work lead by Vinnie @vinnieviruses.bsky.social and in collaboration with @vijayrathinam.bsky.social in @nature.com www.nature.com/articles/s41...
06.08.2025 15:36 — 👍 81 🔁 40 💬 7 📌 2

“Revolutionary Science Comes from Unexpected Angles”
Thoughts from our own Tom Rapoport on the role of basic science in curing disease. magazine.hms.harvard.edu/articles/rev...
29.07.2025 16:00 — 👍 40 🔁 27 💬 0 📌 1

Congratulations to our own @sophiero.bsky.social and team for achieving 3rd place 🥉 in the Merck Innovation Cup 2025. The entire lab is very proud of Sophie's achievement!
25.07.2025 18:00 — 👍 5 🔁 1 💬 0 📌 0
Reactor - The complex formation calculator
No installation required—access Reactor now at reactor-farnunglab.pythonanywhere.com and accelerate your experimental calculation workflows.
22.07.2025 13:18 — 👍 1 🔁 0 💬 0 📌 0
Reactor lets you save, load, import, and export experimental setups (aka recipes); choose between direct recipe formulation or X‑fold compensation modes; and validates inputs in real time to guarantee accuracy and reproducibility of your biochemical experiments.
22.07.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0
Reactor - The complex formation calculator
Here's the solution: Reactor, a web‑based calculator that automates buffer composition and volume calculations for multi‑component reactions: reactor-farnunglab.pythonanywhere.com
22.07.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0
In the Farnung lab, we perform protein complex formation experiments with tens of protein, nucleic acid, and small molecule components. Because of this complexity, calculations to match buffer concentrations often become extremely complicated and tedious. 2/n
22.07.2025 13:13 — 👍 0 🔁 0 💬 1 📌 0
Reactor - The complex formation calculator
I am pleased to announce Reactor—the complex formation calculator for multi-component protein complex formation experiments. Developed by the Farnung Lab @harvardmed.bsky.social.
reactor-farnunglab.pythonanywhere.com 1/n
22.07.2025 13:13 — 👍 23 🔁 9 💬 1 📌 0
Beautiful, Kelly. You and your team are on 🔥!
10.07.2025 23:02 — 👍 1 🔁 0 💬 0 📌 0
Asking for a friend, what is the gold-standard™️ for generating a composite cryo-EM map atm?
07.07.2025 14:48 — 👍 0 🔁 0 💬 0 📌 0
Login • Instagram
Welcome back to Instagram. Sign in to check out what your friends, family & interests have been capturing & sharing around the world.
🎥 Our @harvard.edu undergrad Yerosen Daba is runningthe Harvard College Instagram page today. Check out his story here: www.instagram.com/stories/harv... And learn what we do in the Farnung lab (and perhaps you get to meet the man, the myth, the legend aka Boltzmann too)! 📸⚡
30.06.2025 17:15 — 👍 5 🔁 0 💬 0 📌 0
Thank you and see you in November!
19.06.2025 19:51 — 👍 1 🔁 0 💬 0 📌 0
Thank you, Vijay.
19.06.2025 18:20 — 👍 1 🔁 0 💬 0 📌 0
Thank you!
19.06.2025 18:19 — 👍 0 🔁 0 💬 0 📌 0
Thank you, Kelly. Yes, we are incredibly lucky.
19.06.2025 15:26 — 👍 1 🔁 0 💬 0 📌 0

The 2025 Freeman Hrabowski Scholars | HHMI
Freeman Hrabowski Scholars are outstanding early career faculty who have the potential to become leaders in their research fields.
Congratulations to our own Lucas Farnung @lucas.farnunglab.com who has been named as one of 30 Freeman Hrabowski Scholars for 2025 by the Howard Hughes Medical Institute (HHMI). So well deserved deserved !! www.hhmi.org/programs/fre...
18.06.2025 15:25 — 👍 28 🔁 6 💬 0 📌 0

First PhD candidate (congrats, Dr. James!) to graduate today and check out this beautiful nucleosome/ocean cake made by PhD candidate Della Syau:
16.06.2025 21:06 — 👍 23 🔁 0 💬 0 📌 0


Congratulations to 🎓 Dr. Allison James 🎉. Allison is the first PhD candidate to graduate from the Farnung Lab!
16.06.2025 18:39 — 👍 14 🔁 0 💬 0 📌 0
My message to Trump’s NIH Director? No one in America wants us to do LESS cancer research.
No one is asking Trump to make it harder to cure Alzheimer's disease.
Yet Trump is cutting all of this NOW and demanding an $18 BILLION cut to NIH next year. Not on my watch.
10.06.2025 18:52 — 👍 681 🔁 211 💬 17 📌 10

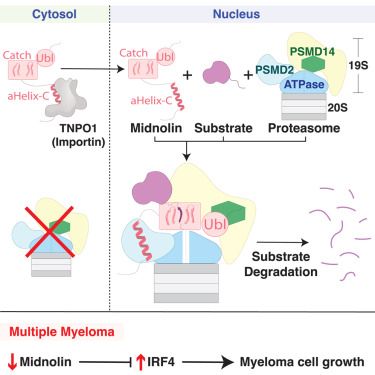
Schulman lab is ready for the GRK2243 Symposium: Understanding ubiquitination: from molecular mechanisms to disease in würzburg
#wUeBI2025 @grk2243.bsky.social
@jakobfarnung.bsky.social @samuelmaiwald.bsky.social @hannahbkmpr.bsky.social
02.06.2025 10:15 — 👍 26 🔁 6 💬 2 📌 0
YouTube video by Fragile Nucleosome
Jon Markert
Check out @jonmarkert.bsky.social's recent talk for the @fnucleosome.bsky.social series, covering his recent breakthrough paper (www.science.org/stoken/autho...) on co-transcriptional histone mark deposition of H3K36me3: www.youtube.com/watch?v=ByJr...
30.05.2025 11:21 — 👍 19 🔁 4 💬 0 📌 0
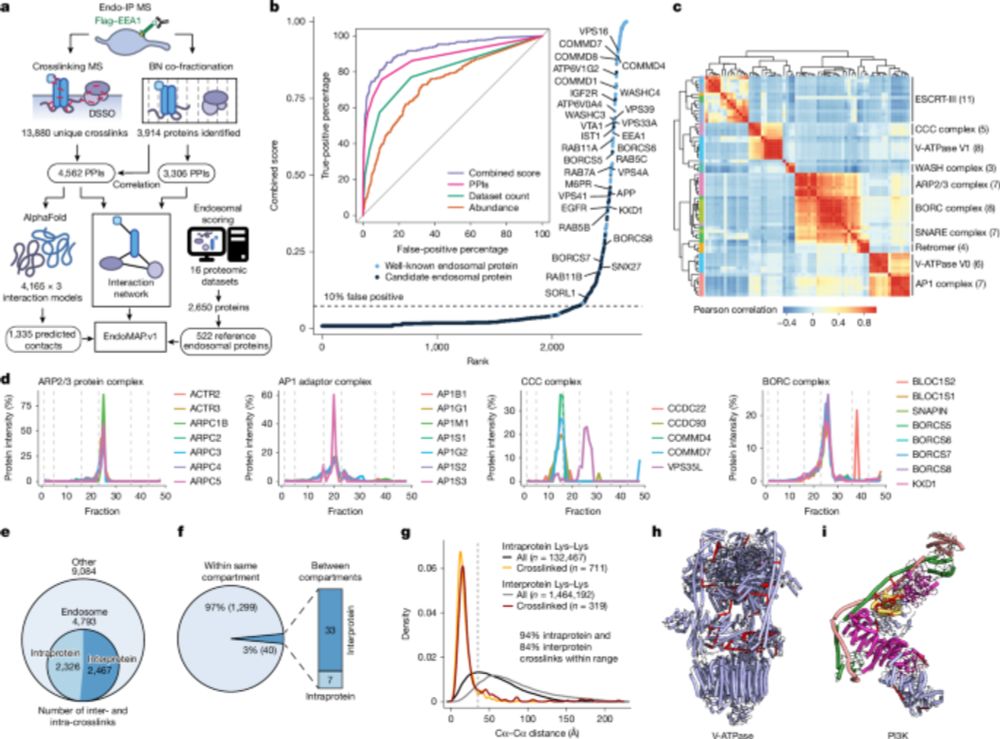
EndoMAP.v1 charts the structural landscape of human early endosome complexes - Nature
A study presents EndoMAP.v1, a resource that combines information on protein interactions and crosslink-supported structural predictions to map the interaction landscape of early endosomes.
New work @harvard by Miguel Gonzalez-Lozano @harperlabhms.bsky.social & @ernstschmid.bsky.social in Johannes Walter lab charts structural interactome of endosomes. #XL-MS #Alphafold Funded by @asapresearch.parkinsonsroadmap.org & NIH. Science continues-despite attacks www.nature.com/articles/s41...
28.05.2025 15:52 — 👍 43 🔁 19 💬 0 📌 1

Farnung Lab
Outstanding work from Allison James who will graduate next month! If you like our work, consider applying to the Farnung Lab. More info: farnunglab.com
06.05.2025 15:09 — 👍 3 🔁 3 💬 0 📌 0

🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
06.05.2025 15:08 — 👍 103 🔁 24 💬 4 📌 3
Watching Sven elucidate this mechanism from an unbiased screen onwards has been one of my favorite experiences as a PI. In these challenging times for science at Harvard, it reminds me of the sheer joy of doing science and making discoveries.
30.04.2025 11:16 — 👍 27 🔁 6 💬 0 📌 0

CONGRATULATIONS!! to @harvardcellbio.bsky.social faculty member Tommy Kirchhausen for his election to the American Academy of Arts and Sciences. So well deserved! www.amacad.org/new-members-...
23.04.2025 19:38 — 👍 8 🔁 1 💬 0 📌 0
Assistant Professor at the University of Hong Kong, loves chromatin structure and protein design.
Associate Prof | Columbia Biology & Zuckerman Institute
Freeman Hrabowski Scholar | HHMI
We study how the skin-brain axis drives somatosensory behaviors
https://www.abdus-saboorlab.com/
Rapid evolutionary dynamics in viruses, cancer and bacteria. Assistant professor at UW Genome Sciences. federlab.github.io
UC Davis |👩🏽🔬 Sensory <—> Motor 💪🏽| Women’s Basketball Fan | Award-winning Children’s Book Author 🧫📚 🧬 | she/her www.thegriffithresearchgroup.com
Assistant Professor at UC Davis | uterus | placenta | women's health | stem cells, uterine injury, & regeneration | Stanford postdoc | UC Berkeley Ph.D. | Harvard A.B.
zhang.faculty.ucdavis.edu
ElisaZhang.org
Laboratory of Biophysical Chemistry of Macromolecules (LCBM) headed by @beatfierz.bsky.social at EPFL | Run by LCBM members
Die DFG ist die größte Forschungsförderorganisation und die Selbstverwaltungsorganisation der Wissenschaft in Deutschland.
www.dfg.de
Impressum: https://www.dfg.de/de/service/kontakt/impressum
Social Media-Netiquette: https://www.dfg.de/de/service/presse
Assistant Professor @ Princeton MolBio |
From transposons to telomeres and back | bicycles and biochemistry
https://ghanim.lab.princeton.edu
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
VP/CSO @HHMI.bsky.social & Head of #VosshallLab @rockefelleruniv Toward Excellent, Inclusive, & Open Science
PhD candidate in the Farnung lab at Harvard Medical School
President of the Max Planck Society since 2023 – dedicated to advancing science for the benefit of society. | @maxplanck.de
🔗 https://www.mpg.de/praesident
🔗 www.mpg.de/imprint
An interdisciplinary research institution that conducts basic research in two key areas of modern biology: Immunobiology & Epigenetics in #Freiburg, Germany
https://www.ie-freiburg.mpg.de
Department of Biological Chemistry and Molecular Pharmacology at HMS. Dedicated to elucidating and visualizing molecular mechanisms in biology and medicine.
Dr. Brian Strahl is a chromatin researcher at UNC Chapel Hill studying how epigenetics contributes to human biology and disease. https://strahl-lab.org
Group Leader @MRC_LMB /Structural Biologist/Biochemist. Amateur baker in free time #RNAworld #spliceosome #telomerase #telomeres #cryoEM #Xraycrystallography
Husband, Father and grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science magazine, Stand Up for Science advisor, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html