
Congratulations to Mike MacCoss on receiving the Donald F. Hunt Distinguished Contribution in Proteomics Award from US HUPO!
us-hupo.org/Distinguishe...

Congratulations to Mike MacCoss on receiving the Donald F. Hunt Distinguished Contribution in Proteomics Award from US HUPO!
us-hupo.org/Distinguishe...

Congratulations to Bill Noble on receiving the 2026 Gil Omenn Computational Proteomics Award from US HUPO!
us-hupo.org/Computationa...

Fantastic project led by @bo-wen.bsky.social. Excited to see the future uses of AI and transfer learning in proteomics. #massspec #proteomics
www.nature.com/articles/s41...

DeepMVP is a deep learning framework to predict PTM sites and variant-induced alterations across 6 common PTMs.
www.nature.com/articles/s41...
The FDR control in the newer versions looks much better. DIA-NN is an excellent tool. We use it a lot in our lab.
25.07.2025 18:48 — 👍 3 🔁 0 💬 0 📌 0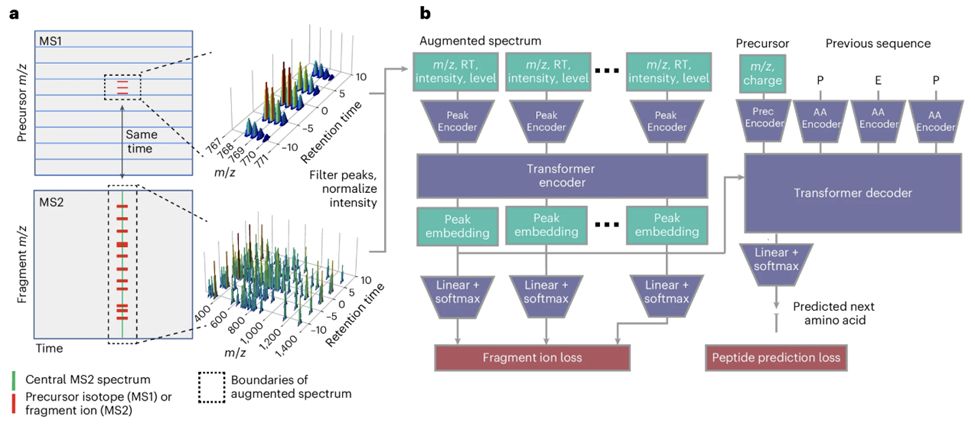
Cascadia from @wnoble.bsky.social is a mass spec-based de novo sequencing model that uses a transformer architecture to handle data-independent acquisition data and achieves substantially improved performance across a range of instruments and experimental protocols. www.nature.com/articles/s41...
07.07.2025 22:31 — 👍 13 🔁 4 💬 0 📌 0The FDR control in the newer version of DIA-NN is much improved on the datasets I have looked at.
04.07.2025 02:42 — 👍 2 🔁 0 💬 0 📌 0(1/2) We have always validated FDR internally on several datasets. Bo Wen and colleagues discovered that FDR of DIA-NN 1.8.1 was anti-conservative on some (but not other) datasets - for some unknown reason. So we fixed it in 2.0 :) Now q-values are more accurate and fluctate less across datasets.
02.07.2025 22:24 — 👍 16 🔁 1 💬 2 📌 0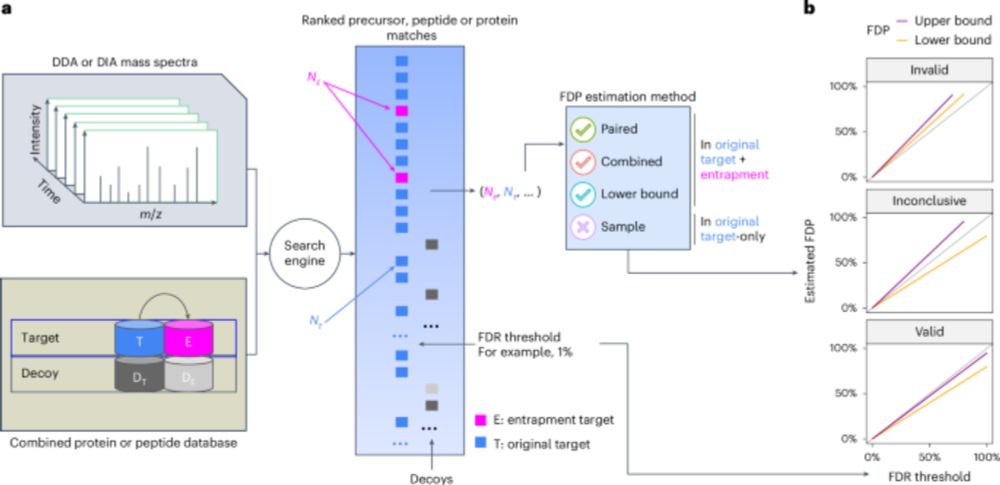
Error control in proteomics mass spectrometry analysis is hard. We came up with a way to evaluate error control. Upshot: for old-school DDA data, not so bad. For DIA data, no existing tool successfully controls the false discovery rate!
www.nature.com/articles/s41...
Excited to see this published! It is a good step in the process for people to assess their FDR control in proteomics experiments. Great work from @bo-wen.bsky.social and @urikeich.bsky.social in particular who drove this.
16.06.2025 18:52 — 👍 42 🔁 9 💬 1 📌 1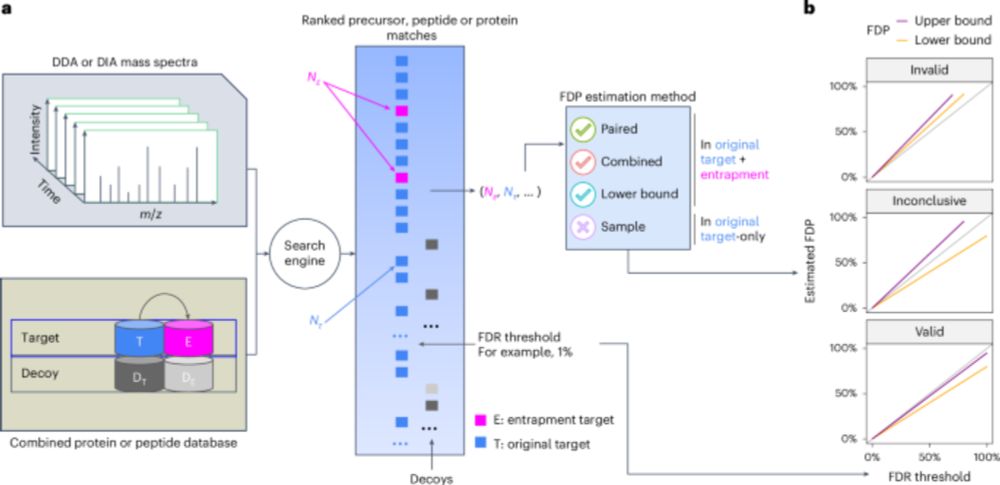
Assessing error control is fundamental in mass spectrometry-based proteomics. @bo-wen.bsky.social @maccoss.bsky.social @urikeich.bsky.social et al introduce a theoretical foundation for entrapment along with a method for more accurate evaluation of FDR control.
www.nature.com/articles/s41...