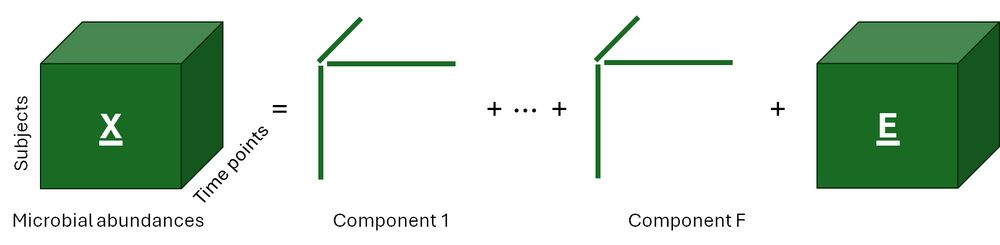Excited to share that my paper on Parallel Factor Analysis for longitudinal microbiome data has been published in mSystems! It details data rearrangement and processing to uncover microbial dynamics. Check it out: buff.ly/5jXEG52
24.05.2025 09:29 — 👍 2 🔁 0 💬 0 📌 0

I'm excited to share that my R package, parafac4microbiome, is updated to version 1.1.2 on CRAN: tinyurl.com/5p78hta3. This release features the full dataset from vanderPloeg2024 (tinyurl.com/y4j58kc8), an improved CORCONDIA, updated vignettes, and much more! #RStats
01.04.2025 14:09 — 👍 5 🔁 1 💬 0 📌 0
I presented my poster today at the Amsterdam Microbiome Expertise Center (AMEC) day at Artis zoo. I'm excited to share it here too. Feel free to message me with any questions! #AMEC2025 #Microbiome #DataAnalysis
13.02.2025 19:29 — 👍 1 🔁 0 💬 0 📌 0
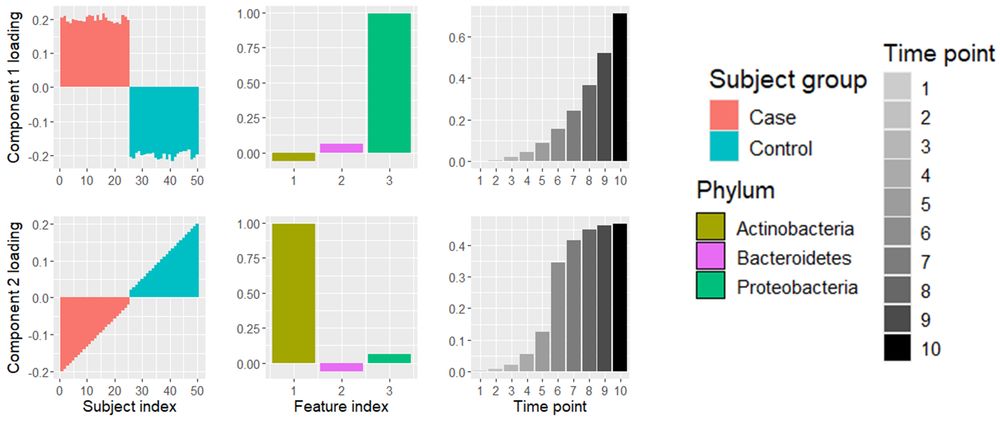
(7/8) PARAFAC reveals distinct microbial growth patterns: Proteobacteria show exponential growth in case subjects, while Actinobacteria exhibit a sigmoid growth curve in controls. This method interprets time-resolved microbial differences between groups more effectively than PCA.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
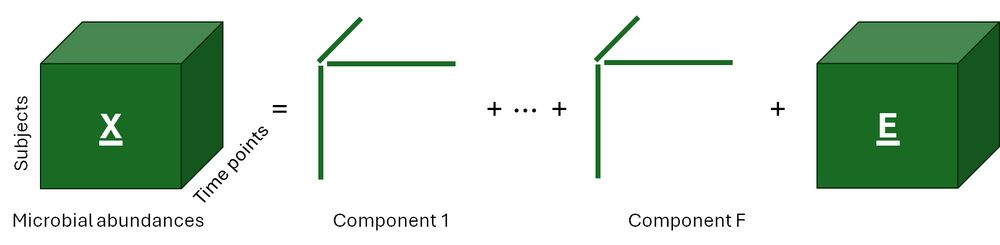
(6/8) In contrast, Parallel factor analysis decomposes a three-way data array into components with three loading vectors each: one for the subjects, one for the microbial relative abundances, and one for time.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
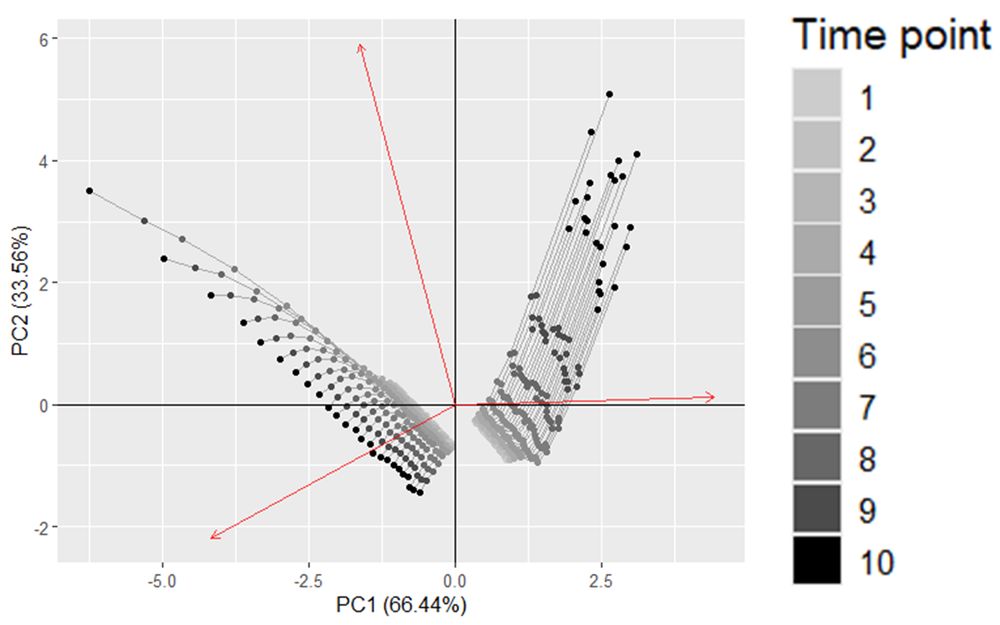
(5/8) The biplot of the PCA shows that the first principal component distinguishes case and control samples by Actinobacteria abundance, while the second separates different time points. However, the time profiles of each subject group remain unclear.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
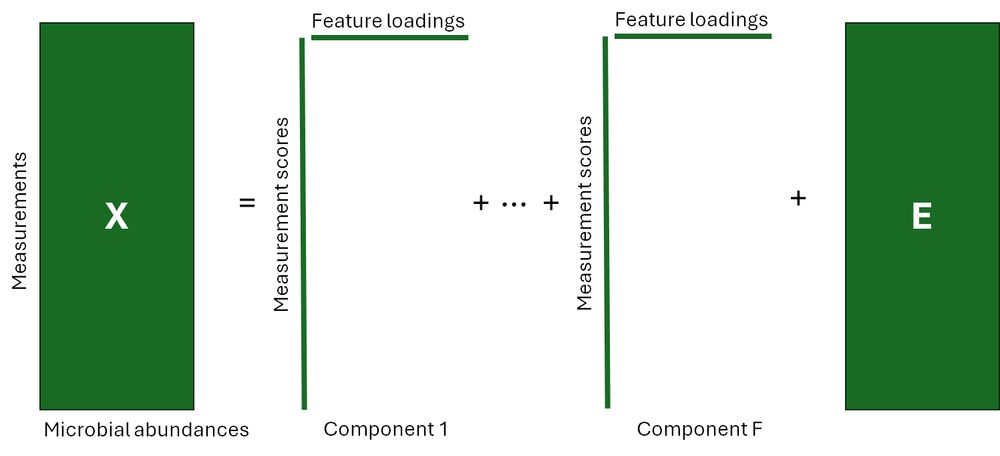
(4/8) Principal Component Analysis would decompose a two-way data table like this into components with scores for the measurements and loadings for the microbial relative abundances. This does not take the longitudinal study design into account.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
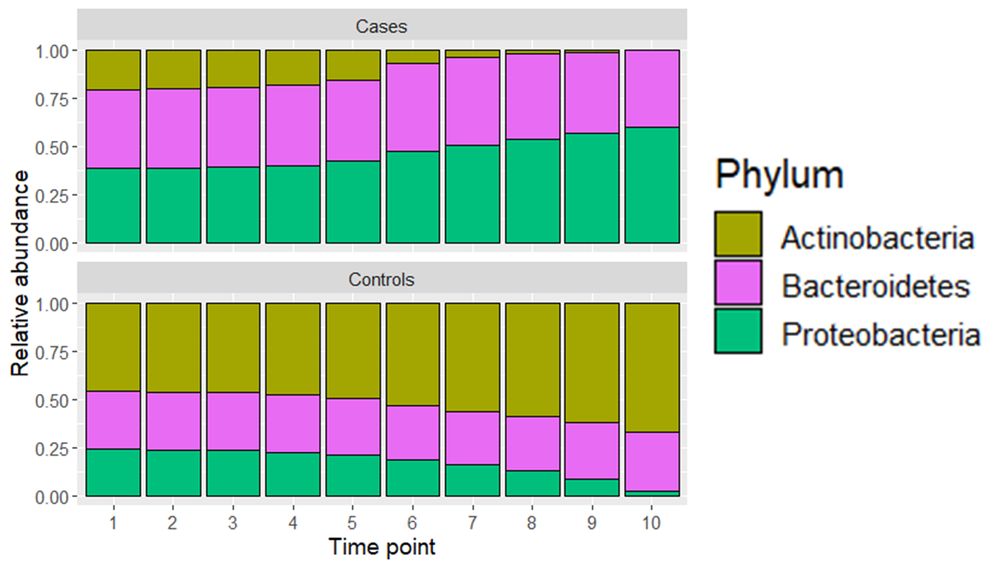
(3/8) The mock data contains three phyla corresponding to 25 case and 25 control subjects across 10 time points. Initially, all subjects are similar. By time point 10, case subjects show reduced Actinobacteria and increased Proteobacteria, while controls show the opposite.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
(2/8) I will highlight how this helps using a a mock intervention dataset, where we have obtained count data of three phyla corresponding to 25 case and 25 control subjects across 10 time points.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0
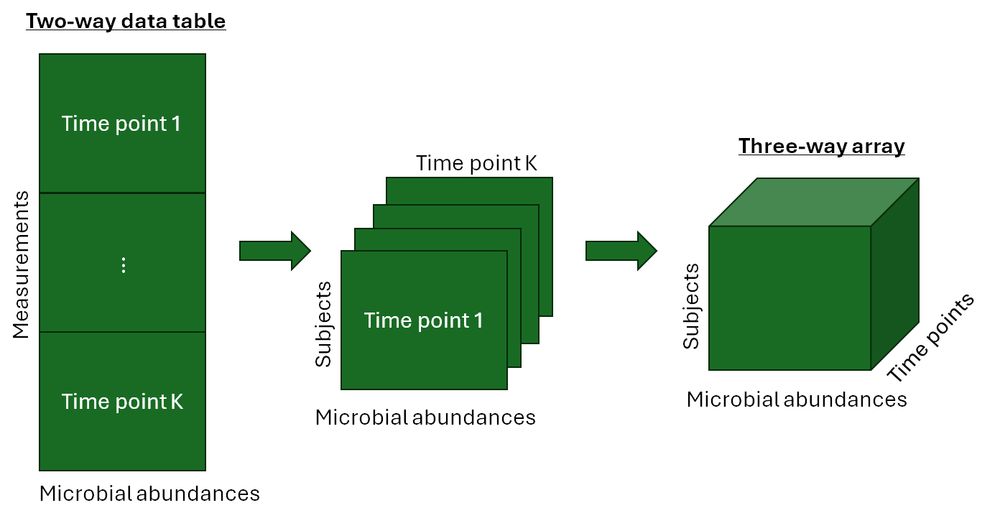
(1/8) Longitudinal microbiome data is shown as a two-way table with rows for the measurements (subject-time point combinations) and columns for microbial abundances, which can be transformed into a three-way array by adding time points as a third dimension.
31.01.2025 19:01 — 👍 0 🔁 0 💬 1 📌 0

Here's a thread 🧵 on using the parafac4microbiome package to track microbial changes with tensors over time. Check out my paper (https://buff.ly/4hDLpFw) and the parafac4microbiome R package (https://buff.ly/4hFVtOD) for more details. #RStats
31.01.2025 19:01 — 👍 2 🔁 1 💬 1 📌 0

G.R. van der Ploeg - About
Happy to share that my personal website grvanderploeg.com is live. You can view my CV, publications and presentations on there if you're interested.
31.12.2024 12:44 — 👍 0 🔁 0 💬 0 📌 0
Happy to be on this platform!
31.12.2024 11:17 — 👍 0 🔁 0 💬 0 📌 0
Postdoc Gut Microbiome & 🫀Health | Nieuwdorp Lab @ Amsterdam UMC | R and Nextflow enthusiast | Hard of hearing 🦻
🔗 https://github.com/barbarahelena
Schetenwapper | MSCA fellow @LabReif @UK_Frankfurt @FfmPsychiatrie | previously @Radboudumc | @tulipantricolor | @NewBrainNutr | 💜microbiota-gut-brain axis
Mostly #rstats in #Ecology, #Nature and #LifeScience
Research scientist at University of Georgia.
Interests: Archaea, human microbiome, anaerobic metabolism, Tungsten enzymes, electron bifurcation.
Onafhankelijke, diepgravende en integere journalistiek. Blijf hier op de hoogte van het laatste nieuws en lees als eerste de nieuwste NRC-artikelen.
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
Oral and gastrointestinal #microbiome #metabolome studies. Topics include relationship to diet and wellness ( #IBD / #cancer / #ostomy/ #ShortBowel syndrome) views are my own.
❤️📊 | 🗣️DE|EN|FR | #rstats | #econsky
Lecturer in Microbiome Science, Blizard Institute, Queen Mary University of London. Studying host-microbiome interactions in early life
🍏Nutritionist
🔬Scientist
🎙️Public speaker
Scientist + Teacher UvA/Amsterdam | Cells | Molecules | Microscopy | Fluorescent Proteins | Biosensors | Lifetime imaging | Open Science | dataViz | R | web apps
Homepage: https://joachimgoedhart.github.io/
DataViz Apps: https://huygens.science.uva.nl
Wnt signaling || Mammary gland || Developmental Stem Cell and Cancer Biology || vanamerongenlab.nl || reneevanamerongen.nl || sils.uva.nl
My Bacon number is lower than my h-index
Create and share social media content anywhere, consistently.
Built with 💙 by a global, remote team.
⬇️ Learn more about Buffer & Bluesky
https://buffer.com/bluesky
microbes, molecules, bioinformatics, ecology @Amsterdam
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app