Congrats! 🎉
07.10.2025 12:27 — 👍 1 🔁 0 💬 0 📌 0Eva D'haene
@eedhaene.bsky.social
postdoc @ http://debaerelab.com @ https://fungenlab-ugent.be | 3D genome, gene regulation, structural variation | retina, neurodev, space!
@eedhaene.bsky.social
postdoc @ http://debaerelab.com @ https://fungenlab-ugent.be | 3D genome, gene regulation, structural variation | retina, neurodev, space!
Congrats! 🎉
07.10.2025 12:27 — 👍 1 🔁 0 💬 0 📌 0Join us today at Sept 23rd2025 from 5pm to 6.30 pm (Central European Time) for our ERN ITHACA webinar on the non-coding genome and human disease by @svergult.bsky.social , @mspielmann.bsky.social , Florence Petit and @stefanbarakat.bsky.social
ern-ithaca.eu/events-news/...

Excited to share the latest work from the lab led by @eharo84.bsky.social, in which we have used synthetic biology to explore the mechanisms by which different types of long-range enhancers ensure robust and precise developmental gene expression
www.biorxiv.org/content/10.1...
👇 Highly recommend applying if you want to join a great team and work on a high-profile, inter-university project! #postdocjobs
31.07.2025 09:46 — 👍 4 🔁 3 💬 0 📌 0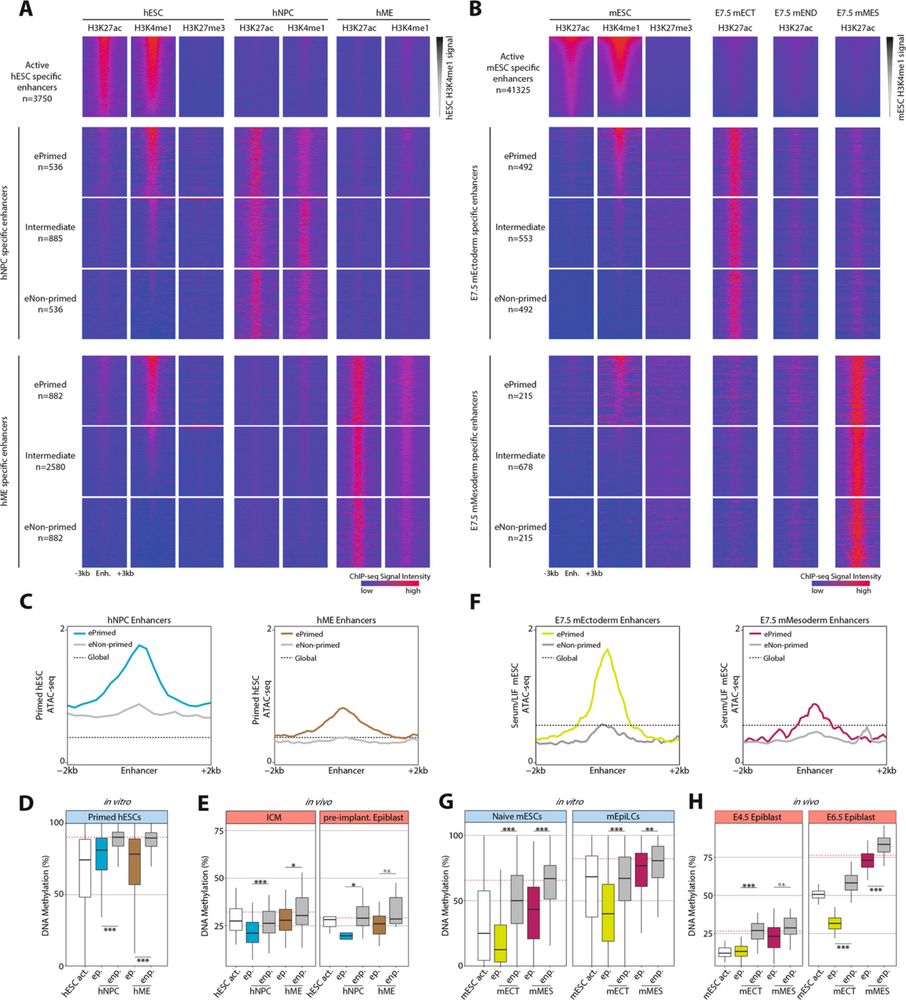
🚨 New paper alert 🚨
I am excited to share our new work on epigenetic enhancer priming in early mammalian development
genomebiology.biomedcentral.com/articles/10....
Fantastic collaboration with Wolf Reik’s team, with key contributions from many others, led by the excellent Chris Todd.
Short 🧵

📣 Join us at the Genetics of Ocular Development (GoOD) Meeting in Ghent, Belgium, 8-9/09/25 🧬
Abstract submission until 16/07/25 goodsoc.org
✨ Exciting line-up ✨
Supported by GoOD @vlaamseoverheid.bsky.social @ds-ugent.bsky.social Lucid
#eyegenetics #development #models #diagnosis #therapy
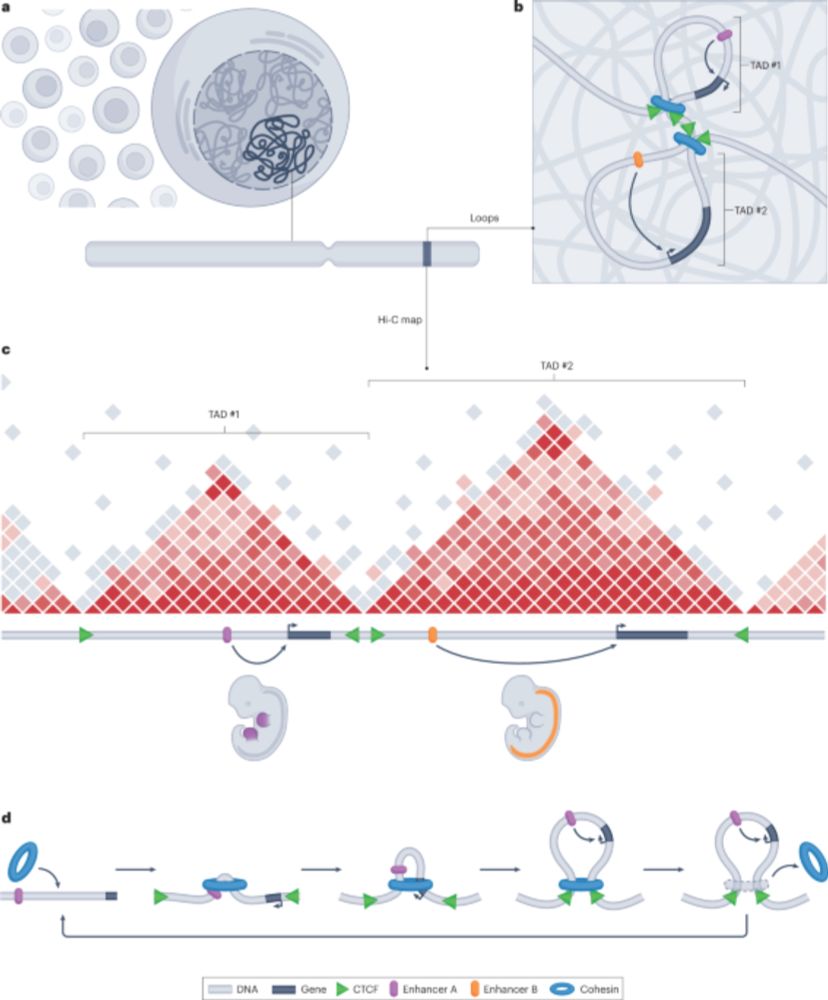
🚀 Thrilled to share our new review on how structural variants reshape 3D genome architecture and cause disease! 🧬🔀
Out now in Nature Reviews Genetics: www.nature.com/articles/s41...
#3D-Genome #StructuralVariants #uksh

Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 — 👍 186 🔁 90 💬 10 📌 9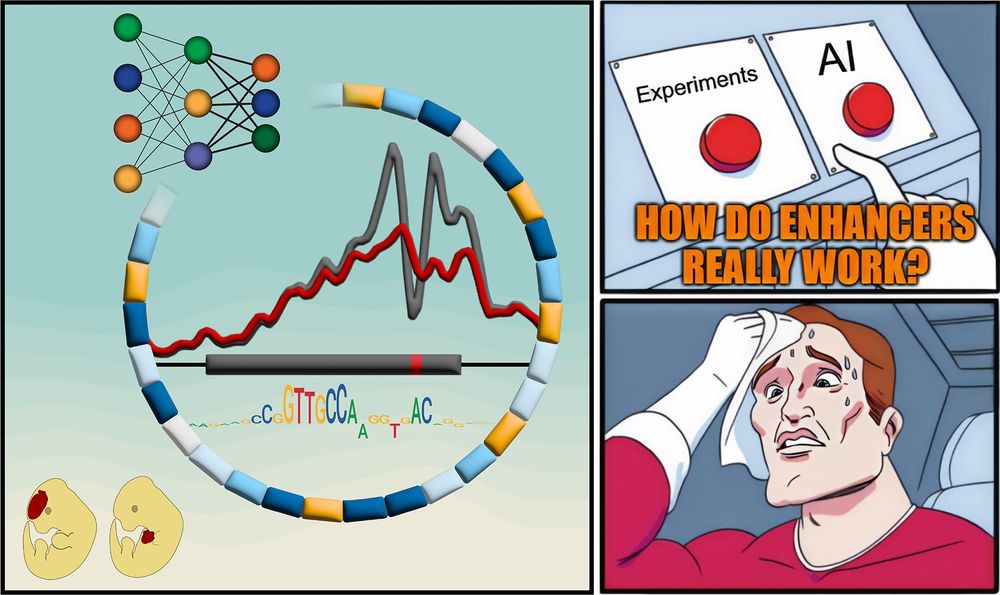
A meme-style comic panel with three parts. Left: A stylized enhancer with a mutation, surrounded by colored blocks representing functional motifs, a neural network diagram, chromatin accessibility signal traces, and a sequence motif. Two cartoon mouse embryos below show different LacZ reporter activity patterns. Top right: A hand hovers anxiously between two red buttons labeled “Experiments” and “AI,” with the caption “HOW DO ENHANCERS REALLY WORK?” Bottom right: A sweating superhero wipes his forehead, looking stressed about the difficult choice.
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22

How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...


Excellent poster talks from all Ghent PhD students at #eshg25! Well done 👏🏻👏🏻
27.05.2025 10:34 — 👍 3 🔁 1 💬 1 📌 0
Excellent talk by my colleague @elfridedebaere.bsky.social on cis regulatory elements in eye diseases at #eshg25. If you did not attend her session, I highly recommend to watch it on the virtual platform. Enhanceropathies research at Ghent University is on 🔥
25.05.2025 16:42 — 👍 8 🔁 2 💬 0 📌 1
@lisahamerlinck.bsky.social kicking ass at #eshg2025
Sharing work on non-coding regulatory disruptions of FOXG1.
#FunGen @svergult.bsky.social @eedhaene.bsky.social
If you're interested and happen to be at #eshg2025, look up @svergult.bsky.social to chat!
25.05.2025 07:13 — 👍 1 🔁 0 💬 0 📌 0We're looking for a new colleague! If you're excited by 2D and 3D neural models for neurodegenerative disease and want to join a fantastic lab, apply!!! 👇
Sharing would be great as well 🙏
#postdoc #jobs #neurogen #neurogenomics
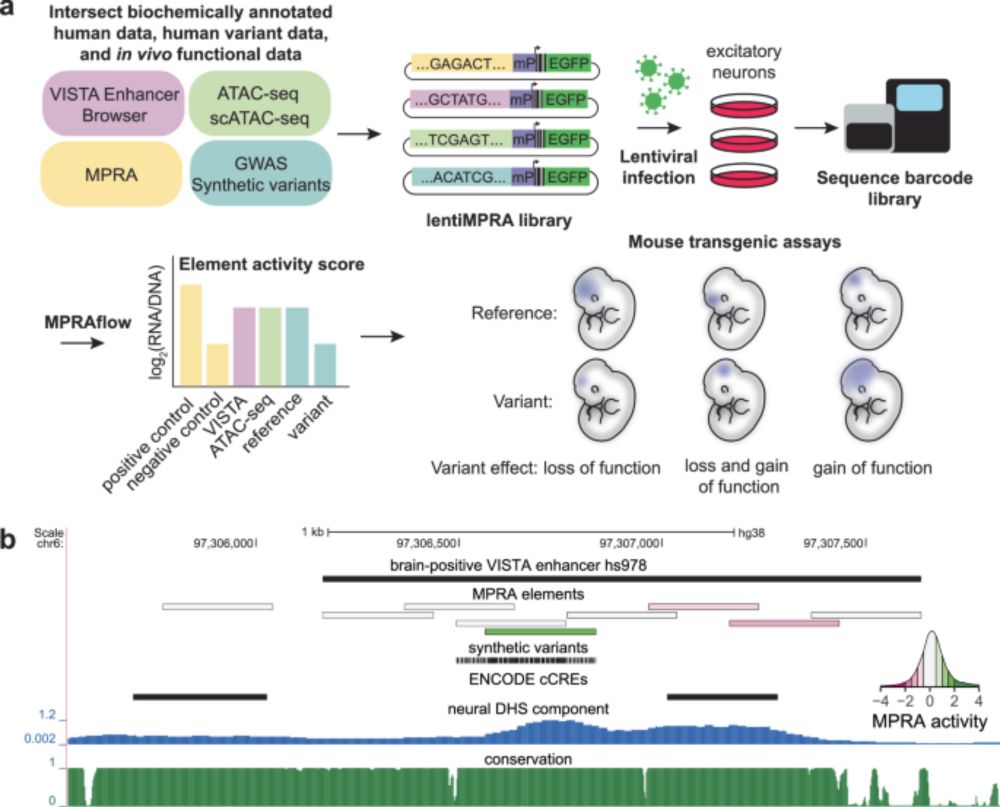
Comparison between a neuronal massively parallel reporter assay for 50,000 sequences and 20,000 variants with mouse enhancer assays provide complimentary information. Great work by Michael Kosicki, Dianne Laboy Cintrón, Len Pennacchio, Martin Kircher lab & many others.
www.nature.com/articles/s41...


On my way to #eshg25!
If you are interested in the work we do at the FunGen lab, check out the flash talk on Sunday and poster on Monday by @lisahamerlinck.bsky.social or come and have a coffee with team members @michael-b-vaughan.bsky.social, @eedhaene.bsky.social or me. Hope to see you there!

🤗 Hugely excited to share our work on automating iterative reanalysis in #raredisease, preprint out: www.medrxiv.org/content/10.1... 🤖🧬
github.com/populationge...
A superb collaboration with @dgmacarthur.bsky.social @cassimons.bsky.social @heidirehm.bsky.social @ksamocha.bsky.social and many more!
Congrats Jasper!
22.05.2025 10:07 — 👍 1 🔁 0 💬 0 📌 0
Looking forward to exciting science and to connect with colleagues in Milan at the Neurogenomics conference @humantechnopole.bsky.social Presenting our research ( @svergult.bsky.social @lisahamerlinck.bsky.social ) on noncoding variants in the FOXG1 locus during the poster session (#88) #neurogen25
19.05.2025 08:39 — 👍 8 🔁 2 💬 0 📌 1Very happy to share the peer-reviewed version of our paper in which we study the formation and function of pair-wise and multi-way enhancer-promoter interactions in gene regulation (see thread below): www.nature.com/articles/s41...
13.05.2025 09:41 — 👍 103 🔁 32 💬 3 📌 3Check out our latest work on the evolution of animal genome regulation out today in @nature.com. Nicely summarized below by @ianakim.bsky.social.
www.nature.com/articles/s41...
This is a major output from our ERC-StG project Evocellmap @erc.europa.eu at @crg.eu

Delighted to share our latest work deciphering the landscape of chromatin accessibility and modeling the DNA sequence syntax rules underlying gene regulation during human fetal development! www.biorxiv.org/content/10.1... Read on for more: 🧵 1/16 #GeneReg 🧬🖥️
03.05.2025 18:27 — 👍 129 🔁 60 💬 2 📌 3
🚨I could not be more excited to share our new preprint on saturation genome editing of the small nuclear RNA (snRNA) RNU4-2:
www.medrxiv.org/content/10.1...
A super fun collaboration with incredible duo @gregfindlay.bsky.social @joachimdejonghe.bsky.social from @crick.ac.uk
🧬🖥️🩺
🧵1/12

Symposium on ‘Enhancer Sequences’ at beautiful @collegedefrance.bsky.social in Paris, by April 11th. Open and free, no registration. Come to relax listening to facts #TherapeuticEffects #VillageGauloix Final programme below. Tell your friends! 🙏🤘
17.03.2025 14:28 — 👍 81 🔁 45 💬 2 📌 1Only one week left to submit your abstract to the @embo.org workshop on "Enhancer mechanics and Enhanceropathies". Submit now and present your latest work in one of our short talks.
10.03.2025 09:06 — 👍 10 🔁 7 💬 0 📌 2Happy to share our review with @akispapantonis.bsky.social in which we summarise and discuss our current understanding of enhancer-mediated gene activation in the context of the 3D genome:
www.annualreviews.org/content/jour...
This multi-year, multi-PhD project was made possible by financial support from the Research Foundation Flanders (FWO) and the Marguerite-Marie Delacroix research fund. @deciphergenomics.bsky.social was instrumental in gathering clinical information.
13.03.2025 16:00 — 👍 3 🔁 0 💬 0 📌 0To sum it up, we identified and characterized regulatory elements essential for proper FOXG1 transcription. Our findings provide new insights into chromatin architecture and gene regulation at the FOXG1 locus, improving SV interpretation in individuals with FOXG1 syndrome-like characteristics.
13.03.2025 16:00 — 👍 1 🔁 1 💬 1 📌 0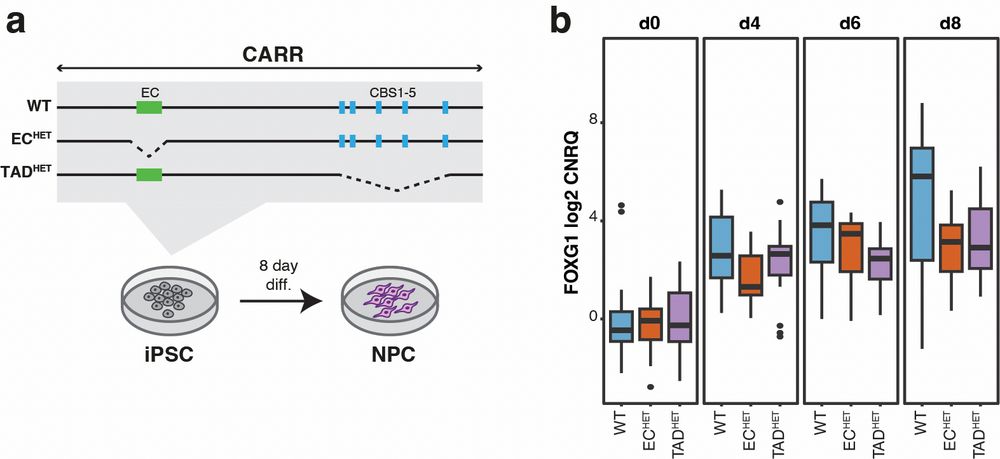
Finally, CRISPR-Cas9 deletion of the enhancer cluster and TAD boundary in NPCs resulted in decreased FOXG1 transcription.
13.03.2025 16:00 — 👍 0 🔁 0 💬 1 📌 0