
Jeg står her omkring i morgen fra ca. kl 10 og frem til 11:30 eller der omkring. Så går turen videre til Panum. Håber vi ses!
16.11.2025 19:53 — 👍 5 🔁 3 💬 1 📌 0
Tusind tak Stinus! Jeg sender informationen ud til så mange som mulig. Jeg undervise dog mellem 9:15 og 12:00. 🫣😤 Først og fremmest, helt og lykke på de sidste meter!
16.11.2025 21:23 — 👍 0 🔁 0 💬 2 📌 0
Tak Stinus. Det ville være fremragende. 100 millioner spørgsmål 😉 strømmer over campus lige nu, som alle prøver at finde svar på. Jeg er sikker på, at et besøg på din gamle hjemmebase ville hjælpe med at finde nogle af disse. 🙏 Ses forhåbentlig mandag.
14.11.2025 06:22 — 👍 1 🔁 0 💬 1 📌 0

Tak for svaret. Definitivt klokken 12. Der er et krydsningspunkt i midten af Nørre Campus (55.7004652, 12.5599248), hvor alle skal forbi mellem bygningerne. Tak! 🙏
13.11.2025 20:08 — 👍 0 🔁 0 💬 1 📌 0
Kommer Du / I også forbi KU, måske endu bedrer Nørre Campus? Jeg tror på Science er der lige i øjeblikket mange med behov for at snakke.
13.11.2025 17:37 — 👍 1 🔁 0 💬 1 📌 0

Diagrams of fruit fly larvae (left) and adult (right) annotated with genes expressed in different tissues, including male- or female-biased gene expressions.

A diagram providing an overview of the regulation of antimicrobial peptide and other host effector peptides by Toll and Imd signalling, alongside diagrams showing the diversity of the peptides.

A summary table illustrating sequence features common to antimicrobial peptides, and a summary table of gene families.

A diagram giving a model for the mechanisms of antimicrobial peptide and host defence peptide actions in killing microbial cells, including precise mechanisms, broader host-pathogen relationships, and non-microbicidal roles of these peptides in protecting the host.
Bizarre that our 2020 #Drosophila #AMP review already feels out of date.
Thanks to #AnnualReviews to cover the many updates in the field and present a much more complete picture of the diversity, mechanisms, topics, of fly immunity re: host defence peptides.
www.annualreviews.org/content/jour...
24.10.2025 12:05 — 👍 29 🔁 12 💬 1 📌 0
We (@sobuelow.bsky.social) developed AF-CALVADOS to integrate AlphaFold and CALVADOS to simulate flexible multidomain proteins at scale
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
20.10.2025 11:26 — 👍 90 🔁 37 💬 0 📌 0
Our collab w. V Goel, @nicholas-aboreden.bsky.social , J Jusuf, G Blobel, L Mirny, @irate-physicist.bsky.social out in @natsmb.nature.com www.nature.com/articles/s41...
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
20.10.2025 18:36 — 👍 55 🔁 18 💬 2 📌 2

Pure bloodlines? Ancestral homelands? DNA science says no. — Harvard Gazette
Geneticist explains recent analyses made possible by tech advances show human history to be one of mixing, movement, displacement.
“Harvard geneticist David Reich said … increasingly sophisticated analysis of genetic material made possible by technological advances shows that virtually everyone came from somewhere else, and everyone’s genetic background shows a mix from different waves of migration that washed over the globe.”
20.09.2025 09:49 — 👍 453 🔁 173 💬 21 📌 19

Guess, I'll have a coffee then.
18.09.2025 10:46 — 👍 0 🔁 0 💬 0 📌 0
Beautiful pictures indeed. What is your weapon of choice: Illustrator or Inkscape?
16.09.2025 09:14 — 👍 1 🔁 0 💬 1 📌 0
Craft House Prague?
15.09.2025 21:18 — 👍 0 🔁 0 💬 0 📌 0
Research Assistant (Fixed Term) - Job Opportunities - University of Cambridge
Research Assistant (Fixed Term) in the Department of Physiology, Development and Neuroscience at the University of Cambridge.
🚨 I’m looking for two enthusiastic scientists to join our team at Cambridge to explore the molecular mechanisms of lactation, early nutrition, and their long-term impact on health.
Apply here:
👉 RA: www.jobs.cam.ac.uk/job/51988/
👉 Postdoc: www.jobs.cam.ac.uk/job/51984/
Please share 🔬👩🔬
24.07.2025 08:34 — 👍 22 🔁 17 💬 0 📌 1
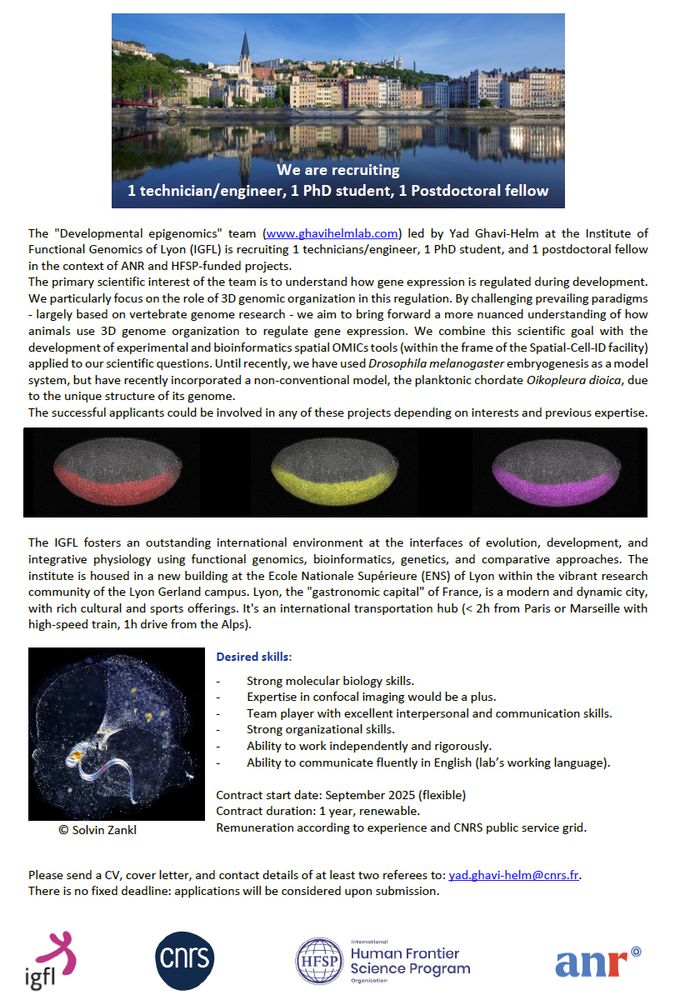
📢 We are recruiting!
Several projects are available and can be tailored to the candidate's profile. Most include confocal imaging/spatial OMICs technologies.
Please share and RT 🙏
23.07.2025 14:37 — 👍 28 🔁 36 💬 1 📌 1

Tomorrow at 19:30 a huge demonstration in Tel Aviv - The Israeli Academia demands to end the war NOW. These horrors are unbearable and must stop.
23.07.2025 15:31 — 👍 111 🔁 24 💬 1 📌 1
“Simple” experiment, but crucial point: we won’t identify all gene regulatory mechanisms underlying disease by simply looking at healthy tissues / cell lines under norm conditions.
19.07.2025 14:16 — 👍 0 🔁 0 💬 0 📌 0
Is this consciously ambiguous? After all he is the Lord of the Dance. 😉
19.07.2025 14:01 — 👍 1 🔁 0 💬 0 📌 0
I second this. Such additional data is great for documentation purposes, but in order for someone (us) to make a systematic change to the publishing model, such stats need to incite a whole field to want to change. As long as authors (us) accept 3 y & ~10 m publishing time, no change will occur.
19.07.2025 13:58 — 👍 3 🔁 0 💬 1 📌 0
I’m the dNTPs and my wife the polymerase. After two cycles, no one cares about where everything started or ends, only that you work together to string a sequence of meaning together.
14.07.2025 20:10 — 👍 8 🔁 0 💬 0 📌 0
So you basically print out the info from the background signal in combination with your actual peak score, so that one could use the former to e.g., color-code the latter based on CNVs?
08.07.2025 14:18 — 👍 0 🔁 0 💬 1 📌 0
Calling all aspiring Postdocs! One extra week to apply to join our exciting HFSP-funded project to uncover how chromatin moves to function.
Deadline July 15th.
www.mdc-berlin.de/career/jobs/...
08.07.2025 09:12 — 👍 15 🔁 16 💬 0 📌 0
GitHub - vierstralab/hotspot3: A chromatin accessibility peak caller with an adaptive background model
A chromatin accessibility peak caller with an adaptive background model - vierstralab/hotspot3
We have created a new DNase I- & ATAC-seq peak caller that uses an adaptive background model that controls for copy number variation & aneuploidy. It performs a per-nucleotide test (+FDR correction) and is very fast. Please try it out and give us feedback!
github.com/vierstralab/...
08.07.2025 03:07 — 👍 41 🔁 10 💬 2 📌 1
Naive question: if you can control for CNVs & aneuploidies, can you so also highlight / annotate peaks identified to be detected in such genome regions. That might be really exciting from a biological point, to contrast peaks inside vs outside?
08.07.2025 09:03 — 👍 0 🔁 0 💬 1 📌 0
Couldn’t agree more, Aarhus is amazing. The university is also visually stunning, great architecture.
07.07.2025 21:55 — 👍 2 🔁 0 💬 0 📌 0
Aarhus centrum, Østergade, I would say. Close to the main shopping street, Søndergade.
07.07.2025 21:39 — 👍 1 🔁 0 💬 1 📌 0
Not stalking you, just know my Berlin. 😉 Half an A, a “l” and a “b” are enough.
07.07.2025 15:12 — 👍 1 🔁 0 💬 1 📌 0
Senior Research Associate, microbiologist, biochemist, The Sorek lab, Weizmann Institute of Science
Researcher @Jiri Lukas lab, Novo Nordisk Center for Protein Research, Genome Instability, CRISPR-Cas9, heritable consequences of DNA repair, single cell high content screening, greenify research 🌱
PhD candidate at MPI-molgen 🧬
Folketingsmedlem @radikale. Ordfører for uddannelse, miljø, udvikling, idræt, medier og erhverv. 📞61624155 / ✉️rvkaro@ft.dk
Diving into genome replication and stability research
Collaborating shoulder to shoulder with Xenopus frogs 🐸 at @bric-ucph.bsky.social, @ucph.bsky.social
Associate Professor at Center for Integrative Genomics (University of Lausanne)
Author of MOTHERLAND: A Feminist History of Modern Russia, from Revolution to Autocracy. A finalist for the National Book Award.
DC correspondent at Puck.
All views my own.
Senior Researcher at the Dep. of GynObs, Hvidovre Hospital. Specialized in women- and reproductive-health.
I leverage a multidisciplinary approach, combining expertise in Bayesian statistics, machine learning, and text mining, with multi-omics data.
Plant molecular biologist at the University of Würzburg
We are the Ting Wu Lab @ HMS. Our laboratory studies how chromosome behavior and positioning influence genome function, with implications for gene regulation, genome stability, and disease.
Managed by WuLab members
https://www.transvection.org/home
💻 🧬 3D genome & RNA lovers Group Leader at Center for Human Technologies, Italian Institute of Technology | CDA @ArmeniseHarvard
Aging and cancer stem cell heterogeneity - ICREA research professor - Quantitative Stem Cell Dynamics lab
at IRB Barcelona 🇦🇷🇪🇺🇺🇸 fraticellilab.com
Associate Professor & Docent @umeauniversitet.bsky.social
Wallenberg Fellow in Molecular Medicine at WCMM Umeå
Postdoc @embl.org
PhD @cniostopcancer.bsky.social
3D genomics , epigenomics, gene regulation, enhancer, cancer, glioblastoma, cancer neuroscience
Chromatin and cancer research at UNC Chapel Hill. Opinions are my own. Raab-lab.org
Postdoc in the Żylicz group at reNEW Copenhagen, formerly in Brand lab at University of Cambridge/NYU Langone;
chromatin, stem cells and metabolism 🔬🧬
Circular RNAs and RNA modifications, Assistant Professor at Yale University
How cells stick to things and move around, microscopy, cats, garden bugs, forays into machine learning, occasional political snark. Asst Prof University of Bath, UK 🏳️🌈
New PI interested in #immune #evolution, host #pathogen interactions, and #ScientificPublishing @ University of Exeter, UK. He/him.
#immunity #infection #antimicrobialpeptides #microbiome #Drosophila #AcademicSky #AcademicChatter #OpenScience 🇨🇦













