
Recording (and most of our past seminars) are released here, www.youtube.com/@CASPRNASIG, enjoy!
04.09.2025 23:28 — 👍 0 🔁 0 💬 0 📌 0@rachael-kretsch.bsky.social
Biophysics PhD Student with DasLab & ChiuLab. Pairing RNA structure & cryoEM to better understand both! King's College, Science & Security '19 Harvey Mudd '18

Recording (and most of our past seminars) are released here, www.youtube.com/@CASPRNASIG, enjoy!
04.09.2025 23:28 — 👍 0 🔁 0 💬 0 📌 0Indeed this seminar will be given by none other than the amazing George Ghanim @automnenine.bsky.social , come learn about how human retrotranposons work and interact with their target DNA
02.09.2025 18:34 — 👍 0 🔁 0 💬 0 📌 0
We have restarted our global Nucleic Acid Strcuture webinar series to bring the expiremental and computational communities together to discuss new developments in the field. Join us this Thursday for the next webinar. Sign up to our mailing list here: groups.google.com/g/casp-rna-sig
02.09.2025 17:50 — 👍 12 🔁 4 💬 1 📌 1When I determed OLE's structure, a transmembrane RNP was staring at me. Took a little bravery to propose this in our manuscript, so I am stoked to see the experts elaborate further. Past data does not rule out OLE RNA spanning the membrane, but data is still needed. Excited for future OLE studies!
04.06.2025 20:25 — 👍 3 🔁 0 💬 0 📌 0And of course thanks to John Moult and Andriy Kryshtafovych for helping recruit all these amazing structural biologists to donate their structures and time to the CASP effort. Thanks for letting me coordinate this paper for the second time, such a cool collection works!
20.05.2025 22:15 — 👍 1 🔁 0 💬 0 📌 0(10) @emillk.bsky.social , Nikolaj H. Zwergius and @esandersen.bsky.social discuss 2’F modified RNA origami and protein-binding aptamer
(11) Heewhan Shin and @phoeberice.bsky.social discuss SPβ LSI-RDF synaptic complex
(12) Reinhard Albrecht, Yimin Hu and Marcus D. Hartmann discuss UDE bound to DNA
(7) Shekhar Jadhav, Michela Nigro, and Marco Marcia discuss group IIC intron
(8) Liu Wang, Jiahao Xie, and Zhaoming Su discuss ROOL RNA nanocage
(9) @rachael-kretsch.bsky.social , Yuan Wu, Rhiju Das, Wah Chiu discuss OLE RNA dimer
(3) Anna Lewicka, Yoshita Srivastava and Joseph A. Piccirilli discuss a 3’ CITE
(4) @cpricejones.bsky.social discusses ZTP riboswitches
(5) Hsuan-Ai Chen and Claudia Höbartner discuss SAMURI
(6) Eric Largy, Philip E. Johnson, and @cdmackereth.bsky.social discuss a DNA aptamer
We each analysed the predictions for the target we expirementally determined, resulting in 12 excellent chapters:
(1) @jgezelle.bsky.social and @lenasteckelberg.bsky.social discuss a xrRNA
(2) Manju Ojha and Deepak Koirala discuss HIV RRE SLII
So much fun working with and learning from some of the best NA structural biologists across the world. It is the hard work of experimental structural biologists that will provide the data and, perhaps most importantly, set the goal-posts for what structure prediction should be aiming to achieve.
20.05.2025 22:15 — 👍 2 🔁 0 💬 1 📌 0
Nucleic acid structural biologists expose that many CASP16 predictions, even ones obtaining high scores by CASP metrics, are inaccurate in the most functionally relevant regions! Read more insights on functionally relevant features by the expert structure determiners (doi.org/10.1101/2025...).
20.05.2025 22:15 — 👍 12 🔁 4 💬 1 📌 1I am excited to see the innovation that will progress nucleic acid structure prediction accuracy as evaluated in RNA puzzles and CASP17! I am perhaps even more excited to see the new ideas that will push our notion of “structure” (or what objectives structure prediction should have) farther.
20.05.2025 21:01 — 👍 2 🔁 0 💬 0 📌 0
CASP16 did challenge predictors with very challenging large RNAs with no previous structures. While no predictors were able to tackle these problems, there are early indications that human experts and automated methods can extract tertiary information from MSAs if sufficiently deep!
20.05.2025 21:01 — 👍 1 🔁 0 💬 1 📌 0
Despite, sub-human performance, servers have seen a jump in performance in CASP16; for the first time servers are able to improve on best available structure template.
20.05.2025 21:01 — 👍 0 🔁 0 💬 1 📌 0The dependence on template availability was observed in RNA-RNA, NA-protein, and NA-ligand interaction prediction in our RNA-multimer, hybrid and NA-ligand category assessments. These categories and the prediction of DNA aptamer structure remain key future challenges.
20.05.2025 21:01 — 👍 0 🔁 0 💬 1 📌 0
Throughout the history of NA structure prediction, accuracy has been dependent on template availability, a trend that unfortunately continues in CASP16. Protein structure prediction has shown this is a surmountable challenge.
20.05.2025 21:01 — 👍 0 🔁 0 💬 1 📌 0Prediction of other motifs, such as A-minor interactions, that form long range interactions important for stitching together the 3D structure remains challenging.
20.05.2025 21:01 — 👍 1 🔁 0 💬 1 📌 0However, CASP16 participants, including automated methods trRosettaRNA and AF3, are more accurate at predicting secondary structure than popular secondary structure prediction algorithms.
20.05.2025 21:01 — 👍 1 🔁 0 💬 1 📌 0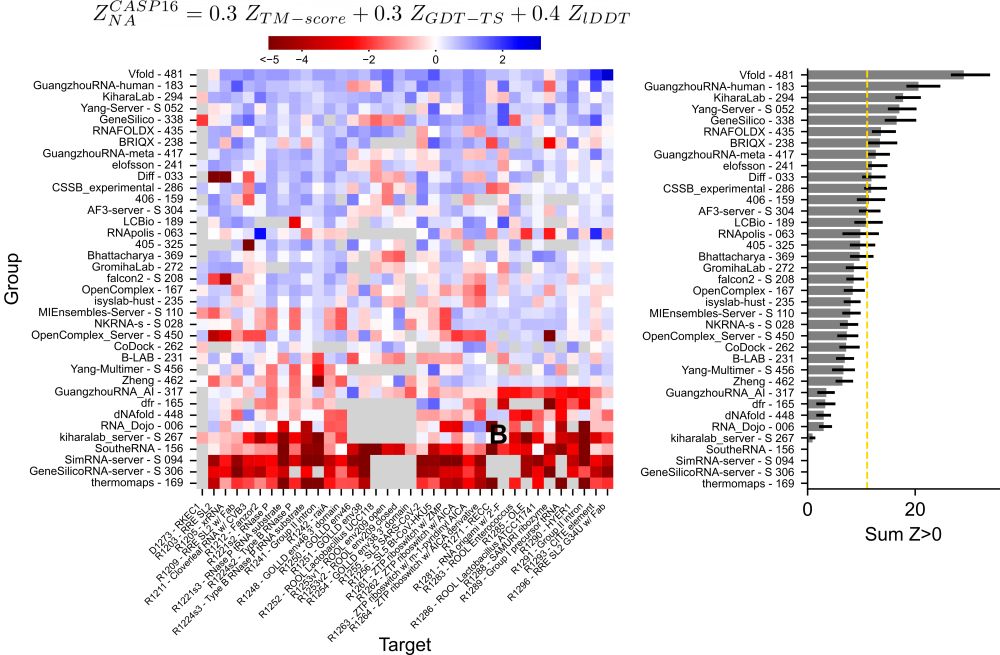
Although the AF3-server can now predict nucleic acid structure, its performance on nucleic acid monomer structure prediction was average; 12 groups out-performed AF3.
20.05.2025 21:01 — 👍 1 🔁 0 💬 1 📌 0
As in CASP15, expert human groups ranked best across targets and categories. These groups are increasingly using deep-learning algorithms to sample structures, which they often then refine or select using physics based algorithms or expert knowledge.
20.05.2025 21:01 — 👍 1 🔁 0 💬 1 📌 0
Work by @alissahummer.com, Shujun He, Rongqing Yuan, Jing Zhang, Thomas Karagianes, Qian Cong, Andriy Kryshtafovych, Rhiju Das, and sizable participation of scientists across the world which provided 42 targets and predictions from 65 groups.
20.05.2025 21:01 — 👍 0 🔁 0 💬 1 📌 0
What is the status of nucleic acid structure prediction? Our analysis of CASP16 (doi.org/10.1101/2025...) reveals human expertise is still necessary for the most accurate prediction, but accuracy still heavily relies on templates; having seen a similar structure already.
20.05.2025 21:01 — 👍 4 🔁 2 💬 1 📌 0Please reach out to me in a message or email if interested!
06.05.2025 01:00 — 👍 0 🔁 0 💬 0 📌 0We will be meeting, Thursdays 3pm UTC = 8am PDT / 11am EDT / 4pm CEST / 11pm CST. For our colleagues in the more difficult timezone, we can accommodate changes in time!
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0We are also looking for excited scientists to help organize! For me, the SIG has been a great way to get to know the RNA community, highly recommended for all early-career scientists interested in the nucleic-acid structure prediction field!
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0We are looking for speaker nominations and self-nominations to discuss work in any of the above fields. If you have read something interesting recently and want to hear more, please forward the information to us!
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0We discuss cool new experimental data describing RNA structure or dynamics, obtaining RNA sequence alignments, predicting RNA or DNA secondary structure, 3D structure, ligand, protein, or RNA–RNA interactions, and ensembles of nucleic acid structures.
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0
We meet virtually, some discussion are recorded on our youtube www.youtube.com/@CASPRNASIG. Join mailing list to keep up to date: groups.google.com/g/casp-rna-sig.
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0We are a group of nucleic acid enthusiasts united by the common goal of producing predictive algorithms for nucleic acid structure prediction.
06.05.2025 01:00 — 👍 0 🔁 0 💬 1 📌 0
With the CASP16 season over (and a defense behind me), I am very excited to say we will be restarting the nucleic acid SIG webinar + discussion series!
We are actively recruited speakers and co-organizers, get in touch!
Read our short piece about the impact of SIG here: doi.org/10.1002/prot...