Protein domains have no single definition, so why stick to one segmentation? 🧩
Instead of forcing structures into rigid classifications, we built AFragmenter. It uses AlphaFold PAE networks for a tuneable approach to domain parsing.
You control the granularity. 👇
🔗 doi.org/10.1093/bioi...
24.11.2025 16:01 — 👍 18 🔁 9 💬 2 📌 0
No worries and valid point. I probably won’t be involved in future efforts but I know that Johannes is definitely looking to expand efforts. Happy to put you two in touch via email if you’d like.
16.11.2025 02:34 — 👍 0 🔁 0 💬 0 📌 0
Thanks! Through the NSF NAIRR pilot program we were paired with an industry partner to support our AI-related project. The program connected us with NVIDIA and they provided three months of access to a 256-GPU DGX cluster, which is where we ran all of the AlphaFold predictions.
14.11.2025 00:10 — 👍 4 🔁 2 💬 0 📌 0
Thanks Andrew!
14.11.2025 00:06 — 👍 1 🔁 0 💬 0 📌 0
Thanks Andreas!
14.11.2025 00:05 — 👍 0 🔁 0 💬 0 📌 0
Thanks Pedro! Our nomination procedure used evidence count from BioGRID as one of the features. We didn’t rely on it (or other dbs) exclusively because we wanted to preserve the ability to nominate interaction pairs that may have been missed by existing physical PPI databases.
14.11.2025 00:05 — 👍 0 🔁 0 💬 1 📌 0
Thanks to members of rhe @johanneswalter.bsky.social lab and our fantastic collaborators @alanbrownhms.bsky.social and Agata Smogorzewska, we experimentally verified several of these new PPIs. But don’t worry, there are plenty more to work on. Come explore the data and see what you find!
12.11.2025 21:26 — 👍 4 🔁 0 💬 1 📌 0
The difference feels like it will be much more informative than overall values.
26.10.2025 17:08 — 👍 2 🔁 0 💬 1 📌 0
End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
22.10.2025 14:30 — 👍 93 🔁 29 💬 2 📌 1

The Farnung Lab celebrates Martin Filipovski's thesis defense.

Martin Filipovski successfully defended his PhD thesis today! Congrats, Dr. Filipovski.
04.09.2025 21:55 — 👍 15 🔁 2 💬 0 📌 0
Just wanted to say that you making this MSA server available to the community is a true service to humanity. Thank you! Have you considered setting up a donation system? I expect many people would contribute!
16.08.2025 16:32 — 👍 5 🔁 0 💬 1 📌 0
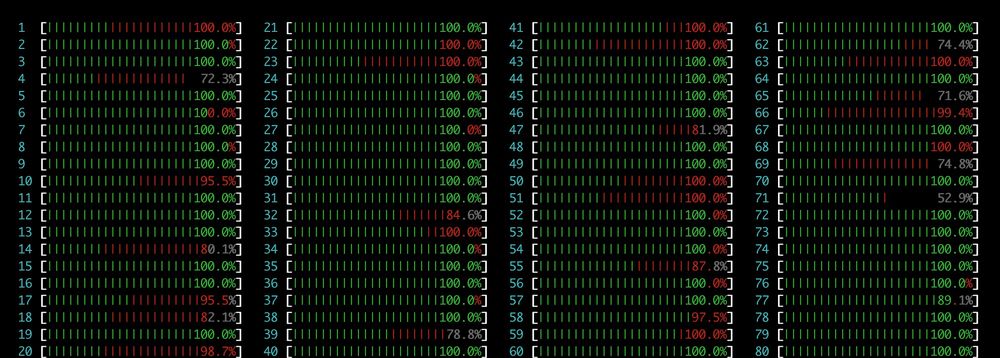

If you use Boltz1/2, BioEmu, Chai1, or other MSA-dependent models, you’re likely using our ColabFold server. Please be considerate! Avoid large submissions across many IPs instead generate the MSA locally. Our server is an old-timer from 2014 and can’t handle that load.
15.08.2025 17:48 — 👍 62 🔁 10 💬 3 📌 0
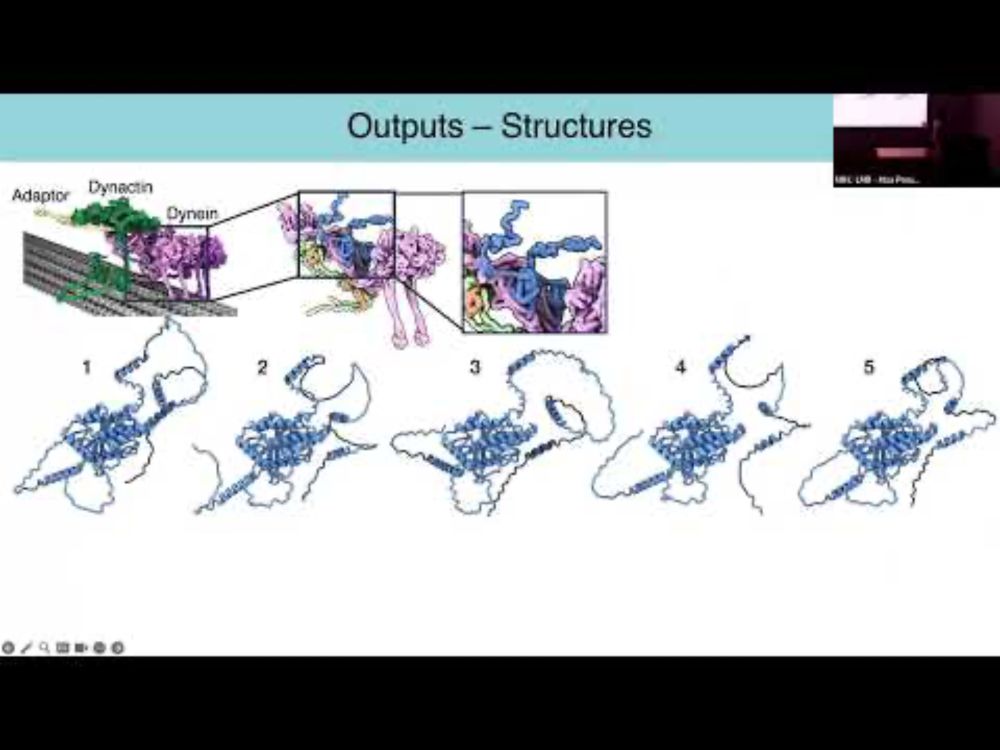
YouTube video by MRC Laboratory of Molecular Biology
Structure Prediction and Design using AlphaFold – Sami Chaaban
Great talk by @sami-c.bsky.social about the proper use and caveats of AlphaFold2/3 predictions, highly recommended to watch www.youtube.com/watch?v=z7vy...
01.08.2025 09:53 — 👍 39 🔁 11 💬 1 📌 0
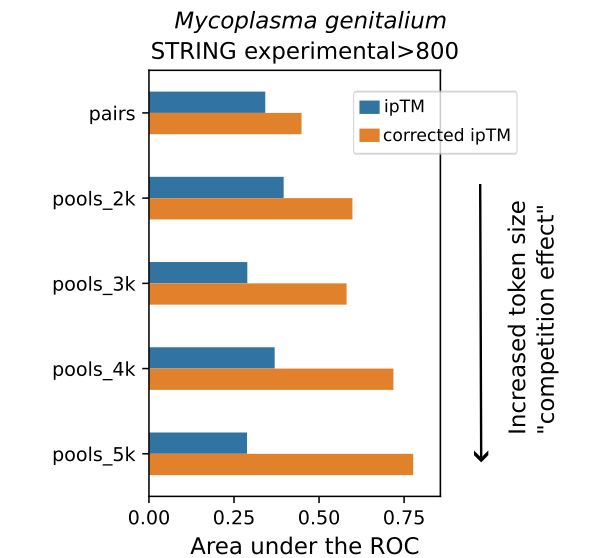
Unexpectedly, @jurgjn.bsky.social found that running Alphafold3 predictions for protein interactions can yield ipTM scores that are more predictive of true interactions when run in pools of proteins instead of pairwise predictions. Presumably, this reflects some sort of "competition effect".
22.07.2025 14:13 — 👍 87 🔁 31 💬 3 📌 3
Folddisco finds similar (dis)continuous 3D motifs in large protein structure databases. Its efficient index enables fast uncharacterized active site annotation, protein conformational state analysis and PPI interface comparison. 1/9🧶🧬
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
07.07.2025 08:21 — 👍 152 🔁 70 💬 8 📌 3

Predictomes | AF3 ANALYSIS
A free interactive resource for viewing predicted AlphaFold multimer structures for protein pairs in biological pathways.
Our AlphaFold 3 prediction analysis tool is now accepting the new version of the AlphaFold 3 server output files. Thanks to those who let me know about the output change and apologies for any inconvenience! predictomes.org/tools/af3/
23.06.2025 14:04 — 👍 3 🔁 0 💬 0 📌 0
Last week I learned that all my NIH awards were terminated due to perceived acts of antisemitism by Harvard. My understanding (which may be incorrect) is that this has been extended to literally every grant to grad students, postdocs and faculty throughout the university.
21.05.2025 01:54 — 👍 0 🔁 3 💬 1 📌 0

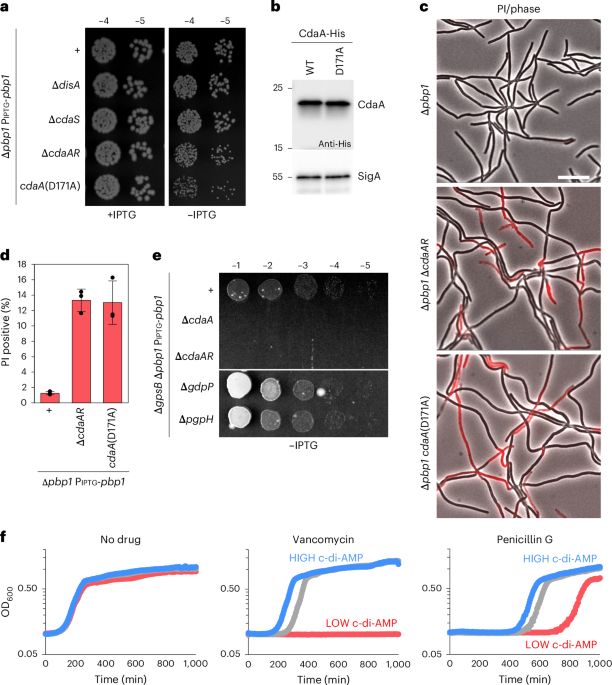
🧬🎉Thrilled to share our new chromatin remodeling study! We reveal three states of human CHD1 and identify a novel "anchor element" that interacts with the acidic patch—conserved among remodelers. Our structures clarify mechanisms of remodeler recruitment! Link: authors.elsevier.com/a/1l2ik3vVUP...
06.05.2025 15:08 — 👍 103 🔁 24 💬 4 📌 3
🔬 New from the Farnung Lab: We established a fully in vitro reconstituted chromatin replication system and report the first cryo-EM snapshots of the human replisome engaging nucleosomes. Brilliant work by @felixsteinruecke.bsky.social with support from @jonmarkert.bsky.social! tinyurl.com/replisome
06.04.2025 09:45 — 👍 179 🔁 57 💬 10 📌 8
Assistant Prof at Utrecht University, studying the process of transcription elongation vs early termination. (she/her)
David Bartel's lab @WhiteheadInst @MIT @HHMI | microRNAs, mRNAs, and other RNAs
Post-doc at Harvard Medical School. Interested in single-molecule Biophysics.
Associate Professor @ Penn Biology, Penn Epigenetics Institute, Penn Center for Genome Integrity.
We combine evolutionary genetics and cell biology to study how DNA repeat evolution perturbs and preserves genome integrity.
Computational chemist/structural bioinformatician working on improving molecular simulation at MRC Laboratory of Molecular Biology. jgreener64.github.io
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
Postdoctoral researcher in the Boulton Lab 🧬 The Francis Crick Institute, London, UK▪️interest in genome stability, chromatin, cancer 🧫 👩🔬 EMBO and MSCA fellow
Our team studies #Rloop & #RNA 🧬 with #biochemistry #cryoEM 🔬❄️ #crystallography💎 to understand how they control life! Views from Charles
https://biochem.emory.edu/bou-nader-lab/
Researcher at Universidad Andres Bello- Chile working on host-bacteria interactions & contact-dependent systems #T3SS and #T6SS. Pew LAF class 2013 and HHMI IRS 2017-2022
Postdoc @ DOE JGI Berkeley Lab 🇺🇸 | Former PhD Student @ Flinders University 🇦🇺 | #bioinformatics #viruses #AI 🦠💻🧬 | she/her
https://github.com/susiegriggo
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Assistant Professor at UMich with a focus on computational pharmacology (he/him)
* Founder and Chief Innovation Officer at Psivant Therapeutics
* Past:
* Chief Computational Scientist at Roivant
* Founding CSO at Silicon Therapeutics
* Global Head, Applications Science at Schrodinger
* Education:
* PhD MIT
* BA & BS UCSB
Postdoc with Sascha Martens and Tim Clausen at the Vienna BioCenter (VBC). #MSCA Fellow. Formerly University of Oxford and University of Antwerp.
Shooting electrons, ions and photons (mainly) at plants to study cell-cell communication @mpibiochem.bsky.social & @hhu.de
Researcher interested in how molecules traverse membranes. Structural biology, biochemistry and biophysics. 🥝
Assistant Professor, Columbia University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Official feed of the Plaschka Lab | Molecular Biology | Research and updates from the team; posts from Clemens are signed –CP.
DNA repair & topoisomerases