🚀 As first official act, we are hiring! 🎓
We’re looking for a PhD student to work at the interface of computational biophysics, machine learning & human mutations. 📌 FPI fellowship, 4 years fully funded!
More information here:
www.bsc.es/join-us/job-...
02.10.2025 08:12 — 👍 15 🔁 9 💬 2 📌 0
Thanks! 🙌
03.10.2025 09:18 — 👍 0 🔁 0 💬 0 📌 0

After months of buildup, it’s finally real! 🎉 The Evolutionary Systems Biophysics Group (ESBG) is officially alive at @bsc-cns.bsky.social . Proud to start this new adventure as a Ramon y Cajal Junior Group Leader🧬
Thanks to all who have been part of this process!
tinyurl.com/4r9vf4zx
02.10.2025 08:06 — 👍 10 🔁 3 💬 0 📌 1
Gracias! 😱
02.10.2025 08:13 — 👍 1 🔁 0 💬 0 📌 0
🚀 As first official act, we are hiring! 🎓
We’re looking for a PhD student to work at the interface of computational biophysics, machine learning & human mutations. 📌 FPI fellowship, 4 years fully funded!
More information here:
www.bsc.es/join-us/job-...
02.10.2025 08:12 — 👍 15 🔁 9 💬 2 📌 0

After months of buildup, it’s finally real! 🎉 The Evolutionary Systems Biophysics Group (ESBG) is officially alive at @bsc-cns.bsky.social . Proud to start this new adventure as a Ramon y Cajal Junior Group Leader🧬
Thanks to all who have been part of this process!
tinyurl.com/4r9vf4zx
02.10.2025 08:06 — 👍 10 🔁 3 💬 0 📌 1
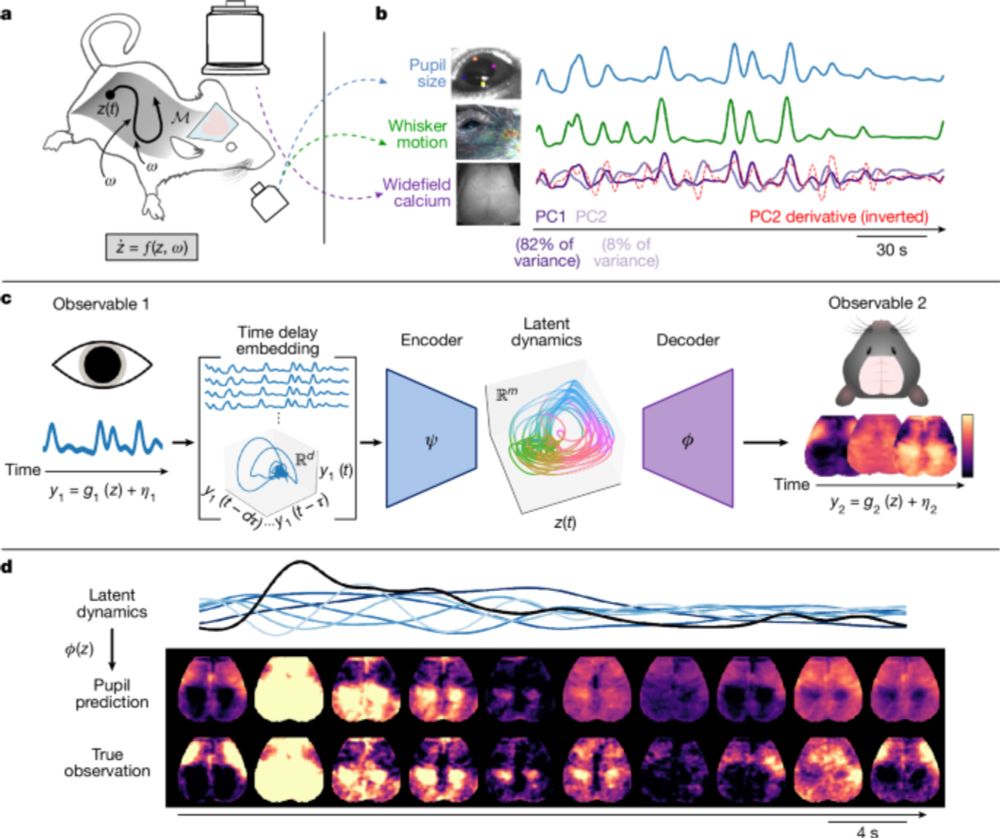
Arousal as a universal embedding for spatiotemporal brain dynamics - Nature
Reframing of arousal as a latent dynamical system can reconstruct multidimensional measurements of large-scale spatiotemporal brain dynamics on the timescale of seconds in mice.
Whole brain dynamics in the ~10s regime shows synchronized propagation of topographically organized rotating waves, driving behavior, calcium fluxes, band-limited power, metabolism, blood flow, etc. All can be predicted from time-lagged embedding of pupil size alone! www.nature.com/articles/s41...
25.09.2025 20:16 — 👍 4 🔁 1 💬 0 📌 0

Delighted to see our paper studying the evolution of plasmids over the last 100 years, now out! Years of work by Adrian Cazares, also Nick Thomson @sangerinstitute.bsky.social - this version much improved over the preprint. Final version should be open access, apols.
Thread 1/n
25.09.2025 21:28 — 👍 298 🔁 153 💬 14 📌 8
LinkedIn just notified about my 4th year annyversary working at the @bsc-cns.bsky.social, time flies!!! It's been a great chapter so far in my scientific journey!! A lot of projects on the list! #ILoveScience
22.09.2025 09:25 — 👍 1 🔁 0 💬 0 📌 0

🥁 Check out our new preprint on OmniPath, the prior knowledge resource for #SystemsBiology, and its brand-new OmniPath Explorer web app! 🥳
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
17.09.2025 13:09 — 👍 33 🔁 18 💬 1 📌 0

Figure 3 from the paper with the caption: "Role of machine learning in de novo design of IDRs. (A) Machine-learning models can be trained on diverse data sources, from molecular dynamics simulations to annotations of cellular localization and protein structures from the Protein Data Bank. (B) Often implemented as neural networks using sequence-encoded features as input, these models can initially be trained on a limited region of sequence space as surrogate models. Through active learning, additional simulations are performed during the design campaign to generate new data, and the surrogate model is retrained on the expanded dataset to progressively improve its accuracy. (C) Machine-learning models have been developed to predict biophysical observables, biological annotations, and protein structures. When combined, machine-learning models can be used to identify a set of sequences that strike a trade-off between multiple design objectives, defining a Pareto front."
New review on computational design of intrinsically disordered proteins 🖥️🍝 by @giuliotesei.bsky.social @fpesce.bsky.social & 👴
doi.org/10.48550/arX...
17.09.2025 16:17 — 👍 48 🔁 16 💬 0 📌 1

10 Rules for a Structural Bioinformatic Analysis
The Protein Data Bank (PDB) is one of the richest open‑source repositories in biology, housing over 277,000 macromolecular structural models alongside much of the experimental data that underpins thes...
Structural bioinformatics is incredibly powerful on its own or when paired with theory or experiment. One of the PDB's superpowers isn’t from one structure, but comparing many to uncover folds, binding sites, and subtle conformational shifts. chemrxiv.org/engage/chemr...
11.09.2025 14:28 — 👍 52 🔁 15 💬 1 📌 2
That is great!!! It is for sure super useful! Thanks a lot!
17.09.2025 18:13 — 👍 2 🔁 0 💬 0 📌 0
Dear Colleagues. We are in need of a molecular dynamics trajectory of a "folding upon binding event" where a protein with an IDR folds into its partner. Do you know of any published and available trajectory that we could use?? Thanksss a lot!
17.09.2025 17:40 — 👍 2 🔁 0 💬 1 📌 0
Rob Finn on MGnify, everything bacteria and functions in different environments
16.09.2025 13:55 — 👍 2 🔁 1 💬 0 📌 0

Now Maria Martín from UniProt is telling us how AI-based tools are shaping the future of one of the key resources for protein sequences and function.
16.09.2025 13:34 — 👍 2 🔁 1 💬 1 📌 0

From structures to sequences, now Alex Bateman and the quest to annotate and classify all proteins!
16.09.2025 13:07 — 👍 2 🔁 1 💬 1 📌 0

Starting our afternoon session with a talk by Sameer Velankar, of PDBe and AFDB fame among other endeavours!
16.09.2025 12:46 — 👍 4 🔁 1 💬 1 📌 0

And now @gonzaparra.bsky.social on his first talk on protein frustration as a PI! Well done!
16.09.2025 11:32 — 👍 6 🔁 2 💬 1 📌 0

David Jones, on novel folds in AFDB and CATH’s founding being celebrated at a now-closed Pizza place in Euston Station
16.09.2025 11:09 — 👍 4 🔁 1 💬 1 📌 0

From CATH to Computational Enzymology, Dame Janet Thornton on the birth of CATH and beyond!
16.09.2025 10:44 — 👍 4 🔁 1 💬 1 📌 0

First Keynote by Burkhard Rost, on the impact of protein language models on the field of structural biology
16.09.2025 10:19 — 👍 4 🔁 1 💬 1 📌 0

Kickstarting our symposium “Protein Annotations in the age of AI” at UCL!
16.09.2025 10:19 — 👍 7 🔁 2 💬 1 📌 0
Stephen Jay Gould was born OTD in 1941.
“Organisms are not billiard balls, propelled by simple and measurable external forces to predictable new positions on life's pool table.... Organisms have a history that constrains their future in myriad, subtle ways.”
🦫🦋🐋🌱🧠🗃️🧪#HistSTM #PhilSci #EvoBio
10.09.2025 16:41 — 👍 158 🔁 44 💬 8 📌 8

Global effort to sequence all life on Earth ramps up
The Earth BioGenome Project is accelerating its efforts tenfold to sequence all life on Earth by 2035.
In the face of climate change, the race to understand life on Earth is on.
@ebpgenome.bsky.social is scaling up tenfold to sequence all known species by 2035.
At EMBL-EBI, we annotate these genomes, making them openly available for the global research community.
www.ebi.ac.uk/about/news/a...
09.09.2025 14:13 — 👍 21 🔁 6 💬 0 📌 1

🚨 New in PNAS!
🧬 64% of disease co-occurrences can be explained by transcriptomic similarities.
Comorbidities aren’t random—they have a molecular basis.
Here’s how we found it 👇 (1/n)
🔗 doi.org/10.1073/pnas.2421060122
@alfonsovalencia.bsky.social
02.09.2025 11:41 — 👍 27 🔁 14 💬 1 📌 4
Postdoc in Andreas Wagner's lab. I'm currently obsessing over how new promoters and enhancers emerge de novo. Also dogs 🐶 and triathlon 🏊 🚲 🏃. He/him 🏳🌈.
timothyfuqua.com
PhD student University College London: machine learning for protein design and structure-based drug discovery
Structural & Systems Biology Lab @ University of Geneva | Exploring protein & proteome assemblies | Structured thoughts, intrinsically disordered views
Cuenta del Consejo Superior de Investigaciones Científicas, el mayor organismo público de investigación de España.
www.csic.es
CABD, Centro Mixto CSIC-UPO-Junta de Andalucía. Unidad de excelencia María de Maeztu (2022-2025) Estudiamos el desarrollo de vertebrados, insectos y nematodos. Y con bacterias, levaduras,organoides y células
🔗 https://www.cabd.es/en/
We work to understand the evolutionary mechanisms that generate biodiversity and to promote its conservation. Joint research centre of @csic.es and @upf.edu.
Evolutionary biologist, Inst. Evol. Biol. (CSIC, Barcelona). Invertebrate genomics, trying to understand how animals colonised land (and also caves). www.metazomics.com
Mexican Historian & Philosopher of Biology • Postdoctoral Fellow at @theramseylab.bsky.social (@clpskuleuven.bsky.social) • Book Reviews Editor for @jgps.bsky.social • #PhilSci #HistSci #philsky • Escribo y edito • https://www.alejandrofabregastejeda.com
Agencia Estatal de Investigación de España. Fomentamos la investigación científica y técnica mediante la asignación de los recursos públicos.
Chief Data Officer at Xyme.
Enzyme Design, Data Science, Bio(Chem)informatics, AI/Machine Learning
Professor of Chemistry, University of Bristol. Computational chemistry,enzyme catalysis, biomolecular simulation,HPC,antibiotic resistance.Views my own.
Professor & Scientist @uspoficial.bsky.social 🇧🇷. Visiting @mit.edu. Scholar @ieausp.bsky.social. Computational (bio)chemistry and biophysics.
Web: http://gaznevada.iq.usp.br
🐦 : x.com/ArantesLab
🎵 : www.dinamicas.art.br
Computational biophysicist | loves lipids and ion channels | bakes, reads, plays board games, runs a little island on animal crossing, collects stamps and stickers, also loves reading about writing
Final year PhD candidate in the Heras Lab at the La Trobe Institute for Molecular Science in Melbourne 🧬 Structural biologist studying the roles of disulphide bond formation proteins in bacteria 🧫
Protein folding nerd. Grad student (at my age?! Yes!) King lab, UW IPD. She/her.
Protein ML. Synbio. Enzyme discovery and engineering. Glandular trichomes. Proud Washington State alum.
Ich habe nichts zu sagen!
Postdoc at University of Leeds #computationalchemistry particularly interested in #molecularmodelling, #ionchannels and #antimicrobialresistance #nanopores
queer (he/him)
YSBL, Department of Chemistry, University of York, UK. I make, modify and fix stuff. Acta Cryst F main editor with Mark and Cristy. CCP4wg2 chair with Arnaud. Analyse glycan structures with Privateer: https://privateer.york.ac.uk #RealTimeChem [He/Him]
PhDing at Barcelona Supercomputing Center | Exploring disease co-occurrences through omics, bioinformatics & HPC — with an eye on AI bias, and occasionally covered in clay.