We’d love to hear ideas for where phosphorylation - or PTM-specific binders could make a difference. If you’re interested in collaborating, please reach out! (8/8)
30.09.2025 21:55 — 👍 0 🔁 0 💬 0 📌 0
This was only possible in a team effort with great people! Thanks @jasonzxzhang.bsky.social, Kejia Wu, Brian Coventry, Gyu Rie Lee, Kody Klupt, Jiuhan Shi, Rafael Brent and the entire @uwproteindesign.bsky.social, with guidance from David Baker and many collaborators. 🙏 (7/8)
30.09.2025 21:55 — 👍 2 🔁 0 💬 1 📌 0

Specificity matters. An all-by-all assay showed strong diagonal binding - each binder recognized only its cognate phosphopeptide, with negligible cross-reactivity. (6/8)
30.09.2025 21:55 — 👍 1 🔁 0 💬 1 📌 0

Crystal structures of CD3ε and EGFR complexes matched design models within ~2 Å. The intended phosphate-binding motifs were reproduced with atomic accuracy, validating the approach. (5/8)
30.09.2025 21:55 — 👍 1 🔁 0 💬 1 📌 0

We tested four sites: CD3ε (TCR), EGFR pY1068 & pY1173, INSR pY1361. In each case, RFD2-MI produced compact proteins that selectively bound the phosphorylated peptide, confirmed by yeast display & BLI with comparable affinities to native phosphorylation binding domains. (4/8)
30.09.2025 21:55 — 👍 1 🔁 0 💬 1 📌 0

We built RFD2-MI, an all-atom diffusion model for molecular interfaces based on RFD2. It co-designs binder + peptide and uses 1D conditioning features like hotspots, secondary structure, and solvent exposure to steer phosphate-pocket formation. (3/8)
30.09.2025 21:55 — 👍 1 🔁 0 💬 1 📌 0
Existing tools fall short: antibodies can’t always tell sites apart, and natural domains like SH2 struggle with specificity. We needed a way to design de novo binders that are both phosphorylation- and sequence-specific. (2/8)
30.09.2025 21:55 — 👍 1 🔁 0 💬 1 📌 0
Phosphorylation on tyrosines control key pathways in immunity, cancer, and metabolism. For the first time, we can now design proteins that specifically recognize individual phosphotyrosines, even in disordered regions. (1/8)
Preprint: www.biorxiv.org/content/10.1...
30.09.2025 21:55 — 👍 44 🔁 18 💬 1 📌 1

Celebrating Seattle's 2024 Nobel Prize with Professor David Baker
YouTube video by UW Medicine
Who knew a Nobel Prize win could unlock an entire city? Join us live on YouTube as we celebrate 2024 Nobel Laureate David Baker together with the Mayor of Seattle and many others on March 10th starting at 5 pm (PT)! 🥇🔑🌇
www.youtube.com/live/z8NO4Bg...
07.03.2025 04:55 — 👍 0 🔁 0 💬 0 📌 0
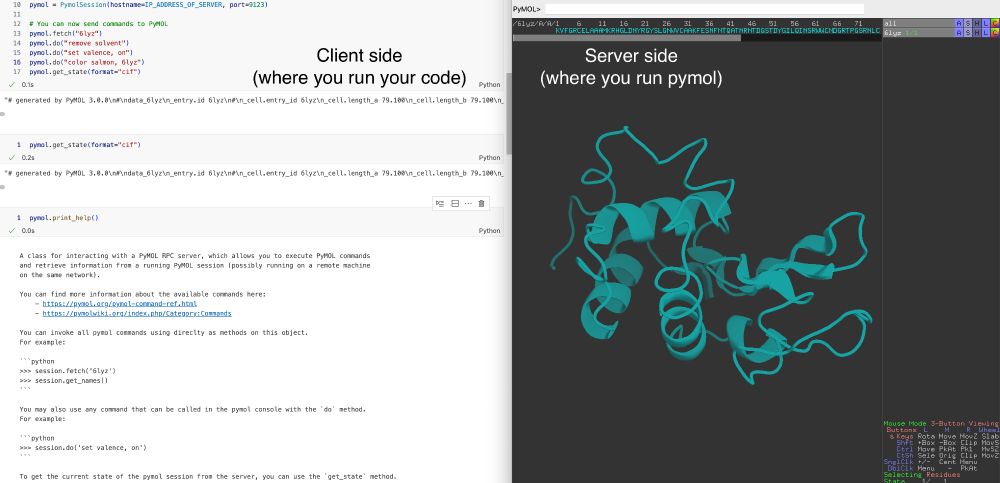
A weekend project from a while back -- this little package (with no dependencies) allows you to interact with pymol remotely.
I use it a lot for my protein design workflows together with @biotite.bsky.social.
Just `pip install pymol-remote`
25.11.2024 14:50 — 👍 47 🔁 13 💬 4 📌 3
#CompChemSky 🧶🖥️🧬🧪
I wonder, is there any computational approach, that would find protein in PDB according to any arbitrary shape?
06.12.2023 19:23 — 👍 3 🔁 2 💬 2 📌 0

Sticker on a laptop saying 'I <3 Biophysics' with all the letters taken form protein structures from the PDB

New sticker design saying 'I <3 Biophysics' with a different heart from SARS-CoV-2 NSP13
I would also be very interested in find some alternative letters, for example a B and P for a new ‘I ❤️ Biophysics’ sticker. Currently its hard to read for some people even with the Howarth and Chroma alphabet. SARS-CoV-2 NSP13 provided an amazing heart btw!
07.12.2023 00:37 — 👍 2 🔁 0 💬 0 📌 0
The Praetorius lab for Biomolecular Design at the Institute of Science and Technology Austria (ISTA) is looking for grad students in 2024. If you are interested in protein design at a great institute near Vienna reach out to me!
www.dropbox.com/scl/fi/6iny2...
28.11.2023 01:08 — 👍 11 🔁 8 💬 1 📌 0
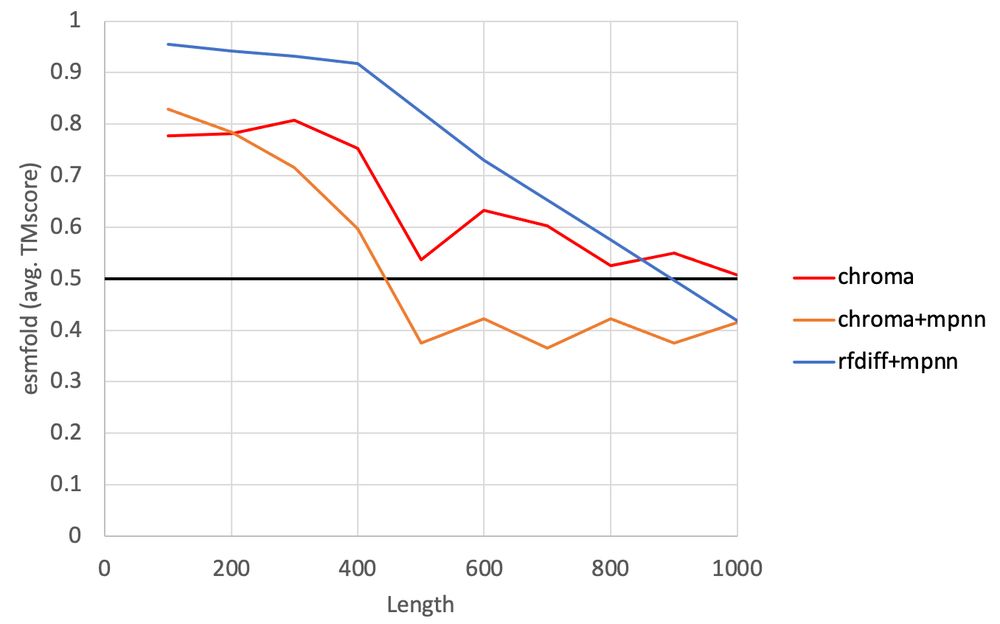
Update on the Chroma vs RfDiffusion analysis.
ProteinMPNN just doesn't like Chroma's backbones (poor prediction of proteinMPNN generated sequences by ESMFold). Interestingly, Chroma's own sequence design method (which was trained in the context of partially noise backbones) loves it! (1/3)
18.11.2023 19:04 — 👍 12 🔁 6 💬 1 📌 1

"Performance and structural coverage of the
latest, in-development AlphaFold model" 🧪🧶🧬
DeepMind & Isomorphic Labs sharing some updates (but no code) on what is presumably alphafold 3, capable of modeling ligands, nucleic acids, antibody-antigen complexes etc
storage.googleapis.com/deepmind-med...
31.10.2023 13:48 — 👍 10 🔁 4 💬 1 📌 1
CryoEM, membrane proteins and whatnot
Assistant Professor at Columbia Chemistry. PI of a chemical biology lab full of awesome people. Internal conflicts: Chemist or biologist? Kinase or phosphatase?
Lab website: https://shahlab.wixsite.com/home
ORCiD: 0000-0002-1186-0626
Computational designer of molecular tools for intracellular signaling. Assistant Professor at UCLA Bioengineering. https://scholar.google.com/citations?user=T_SIIAsAAAAJ&hl=en
Biochemist, Protein Engineer
PhD student Winter lab @ AITHYRA/ CeMM Vienna | interested in using structural and synthetic biology to engineer cellular decision making | previously Baker lab @ IPD and Taylor lab @ mpiib-berlin
Assistant Professor at @EPFL in Computer Science and Life Sciences. PostDoc at @Genentech and @Stanford. PhD at @ETH and @BroadInstitute.
www.aimm.epfl.ch
Investigating protein nanomachines and their dynamics.
ARC DECRA Fellow at Monash University 🇦🇺.
Structural biologist. Author of WIGGLE. He/Him. 🏳️🌈
Scientist, artist, educator, chef, fisherman, outdoor enthusiast, bookworm, linguist, peacemaker, volunteer, etc. in no particular order.
Please don't ask because I will not date you, I will not send you money, and I will not be your disciple. Thanks.
I talk about computational #openscience communication and science publishing. On the jupyterbook.org & @mystmd.org teams, co-founder of @curvenote.com & @continuous.foundation.
PhD Candidate | EPFL
Structural bioinformatics
https://parthbibekar.github.io
ML Scientist @ ENPICOM B.V. (Den Bosch, Netherlands)
computational biology, ML, protein design, cheminformatics, fancy dev tooling, tinge of bouldering
https://marinegor.dev
I’m a fly-fishing virologist with 5 kids, and twin grandsons. I have a passion for viruses, vaccines, and antivirals. And for food, travel, and sailing. And democracy too!
Assistant Professor, University of Toronto Scarborough.
Genetics, plant development, epigenetics, climate change, science fiction and history.
https://www.satyaki-lab.com/
https://scholar.google.com/citations?user=tPfWn1MAAAAJ&hl=en
Senior Research Scientist in comp bio at @deepgenomics. Did PhD work on cancer evolution. My three favourite things are bananas, riding my bike very fast, and vim.
Postdoc @Yale. Incoming Fellow @RTI, UMass Chan
https://schaerfenlab.com/
Protein structure-function | RNA | Infectious diseases | drug discovery
PI in molecular and structural biology
Go!RNA lab, University of Warsaw
https://gorna.uw.edu.pl
https://x.com/m_gorna
ICREA Research Professor at IRB Barcelona. Working on computational cancer genomics. Leading @bbglab.bsky.social
http://bbglab.irbbarcelona.org
Leading the Experimental Plant Systems Biology Lab at KU Leuven 🇧🇪. Previously at UAlberta 🇨🇦, ETH_Zurich🇨🇭, Imperial College London 🇬🇧, VIT University 🇮🇳.
Fmr Early Career Advisor @eLife, @biorxiv affiliate
he/him
mehta-lab.com
Community builder, open source, PBL, math, keynote speaker, public libraries, ukuleles, #EduSky, maker, folk music, digital inclusion. Library Journal Mover & Shaker 2024. https://opensource.com/users/pshapiro
Assistant Professor in Biocatalysis at TU Delft.











